Repository: spark Updated Branches: refs/heads/master 2a29a60da -> 1e44dd004
[SPARK-3181][ML] Implement huber loss for LinearRegression. ## What changes were proposed in this pull request? MLlib ```LinearRegression``` supports _huber_ loss addition to _leastSquares_ loss. The huber loss objective function is: 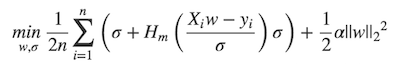 Refer Eq.(6) and Eq.(8) in [A robust hybrid of lasso and ridge regression](http://statweb.stanford.edu/~owen/reports/hhu.pdf). This objective is jointly convex as a function of (w, Ï) â R Ã (0,â), we can use L-BFGS-B to solve it. The current implementation is a straight forward porting for Python scikit-learn [```HuberRegressor```](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.HuberRegressor.html). There are some differences: * We use mean loss (```lossSum/weightSum```), but sklearn uses total loss (```lossSum```). * We multiply the loss function and L2 regularization by 1/2. It does not affect the result if we multiply the whole formula by a factor, we just keep consistent with _leastSquares_ loss. So if fitting w/o regularization, MLlib and sklearn produce the same output. If fitting w/ regularization, MLlib should set ```regParam``` divide by the number of instances to match the output of sklearn. ## How was this patch tested? Unit tests. Author: Yanbo Liang <yblia...@gmail.com> Closes #19020 from yanboliang/spark-3181. Project: http://git-wip-us.apache.org/repos/asf/spark/repo Commit: http://git-wip-us.apache.org/repos/asf/spark/commit/1e44dd00 Tree: http://git-wip-us.apache.org/repos/asf/spark/tree/1e44dd00 Diff: http://git-wip-us.apache.org/repos/asf/spark/diff/1e44dd00 Branch: refs/heads/master Commit: 1e44dd004425040912f2cf16362d2c13f12e1689 Parents: 2a29a60 Author: Yanbo Liang <yblia...@gmail.com> Authored: Wed Dec 13 21:19:14 2017 -0800 Committer: Yanbo Liang <yblia...@gmail.com> Committed: Wed Dec 13 21:19:14 2017 -0800 ---------------------------------------------------------------------- .../ml/optim/aggregator/HuberAggregator.scala | 150 ++++++++++ .../ml/param/shared/SharedParamsCodeGen.scala | 3 +- .../spark/ml/param/shared/sharedParams.scala | 17 ++ .../spark/ml/regression/LinearRegression.scala | 299 +++++++++++++++---- .../optim/aggregator/HuberAggregatorSuite.scala | 170 +++++++++++ .../ml/regression/LinearRegressionSuite.scala | 244 ++++++++++++++- project/MimaExcludes.scala | 5 + 7 files changed, 823 insertions(+), 65 deletions(-) ---------------------------------------------------------------------- http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala ---------------------------------------------------------------------- diff --git a/mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala b/mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala new file mode 100644 index 0000000..13f64d2 --- /dev/null +++ b/mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala @@ -0,0 +1,150 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.spark.ml.optim.aggregator + +import org.apache.spark.broadcast.Broadcast +import org.apache.spark.ml.feature.Instance +import org.apache.spark.ml.linalg.Vector + +/** + * HuberAggregator computes the gradient and loss for a huber loss function, + * as used in robust regression for samples in sparse or dense vector in an online fashion. + * + * The huber loss function based on: + * <a href="http://statweb.stanford.edu/~owen/reports/hhu.pdf";>Art B. Owen (2006), + * A robust hybrid of lasso and ridge regression</a>. + * + * Two HuberAggregator can be merged together to have a summary of loss and gradient of + * the corresponding joint dataset. + * + * The huber loss function is given by + * + * <blockquote> + * $$ + * \begin{align} + * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma + + * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) + \frac{1}{2}\lambda {||w||_2}^2} + * \end{align} + * $$ + * </blockquote> + * + * where + * + * <blockquote> + * $$ + * \begin{align} + * H_m(z) = \begin{cases} + * z^2, & \text {if } |z| < \epsilon, \\ + * 2\epsilon|z| - \epsilon^2, & \text{otherwise} + * \end{cases} + * \end{align} + * $$ + * </blockquote> + * + * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95% statistical efficiency + * for normally distributed data. Please refer to chapter 2 of + * <a href="http://statweb.stanford.edu/~owen/reports/hhu.pdf";> + * A robust hybrid of lasso and ridge regression</a> for more detail. + * + * @param fitIntercept Whether to fit an intercept term. + * @param epsilon The shape parameter to control the amount of robustness. + * @param bcFeaturesStd The broadcast standard deviation values of the features. + * @param bcParameters including three parts: the regression coefficients corresponding + * to the features, the intercept (if fitIntercept is ture) + * and the scale parameter (sigma). + */ +private[ml] class HuberAggregator( + fitIntercept: Boolean, + epsilon: Double, + bcFeaturesStd: Broadcast[Array[Double]])(bcParameters: Broadcast[Vector]) + extends DifferentiableLossAggregator[Instance, HuberAggregator] { + + protected override val dim: Int = bcParameters.value.size + private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1 + private val sigma: Double = bcParameters.value(dim - 1) + private val intercept: Double = if (fitIntercept) { + bcParameters.value(dim - 2) + } else { + 0.0 + } + + /** + * Add a new training instance to this HuberAggregator, and update the loss and gradient + * of the objective function. + * + * @param instance The instance of data point to be added. + * @return This HuberAggregator object. + */ + def add(instance: Instance): HuberAggregator = { + instance match { case Instance(label, weight, features) => + require(numFeatures == features.size, s"Dimensions mismatch when adding new sample." + + s" Expecting $numFeatures but got ${features.size}.") + require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0") + + if (weight == 0.0) return this + val localFeaturesStd = bcFeaturesStd.value + val localCoefficients = bcParameters.value.toArray.slice(0, numFeatures) + val localGradientSumArray = gradientSumArray + + val margin = { + var sum = 0.0 + features.foreachActive { (index, value) => + if (localFeaturesStd(index) != 0.0 && value != 0.0) { + sum += localCoefficients(index) * (value / localFeaturesStd(index)) + } + } + if (fitIntercept) sum += intercept + sum + } + val linearLoss = label - margin + + if (math.abs(linearLoss) <= sigma * epsilon) { + lossSum += 0.5 * weight * (sigma + math.pow(linearLoss, 2.0) / sigma) + val linearLossDivSigma = linearLoss / sigma + + features.foreachActive { (index, value) => + if (localFeaturesStd(index) != 0.0 && value != 0.0) { + localGradientSumArray(index) += + -1.0 * weight * linearLossDivSigma * (value / localFeaturesStd(index)) + } + } + if (fitIntercept) { + localGradientSumArray(dim - 2) += -1.0 * weight * linearLossDivSigma + } + localGradientSumArray(dim - 1) += 0.5 * weight * (1.0 - math.pow(linearLossDivSigma, 2.0)) + } else { + val sign = if (linearLoss >= 0) -1.0 else 1.0 + lossSum += 0.5 * weight * + (sigma + 2.0 * epsilon * math.abs(linearLoss) - sigma * epsilon * epsilon) + + features.foreachActive { (index, value) => + if (localFeaturesStd(index) != 0.0 && value != 0.0) { + localGradientSumArray(index) += + weight * sign * epsilon * (value / localFeaturesStd(index)) + } + } + if (fitIntercept) { + localGradientSumArray(dim - 2) += weight * sign * epsilon + } + localGradientSumArray(dim - 1) += 0.5 * weight * (1.0 - epsilon * epsilon) + } + + weightSum += weight + this + } + } +} http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/main/scala/org/apache/spark/ml/param/shared/SharedParamsCodeGen.scala ---------------------------------------------------------------------- diff --git a/mllib/src/main/scala/org/apache/spark/ml/param/shared/SharedParamsCodeGen.scala b/mllib/src/main/scala/org/apache/spark/ml/param/shared/SharedParamsCodeGen.scala index a267bbc..a5d57a1 100644 --- a/mllib/src/main/scala/org/apache/spark/ml/param/shared/SharedParamsCodeGen.scala +++ b/mllib/src/main/scala/org/apache/spark/ml/param/shared/SharedParamsCodeGen.scala @@ -88,7 +88,8 @@ private[shared] object SharedParamsCodeGen { "during tuning. If set to false, then only the single best sub-model will be available " + "after fitting. If set to true, then all sub-models will be available. Warning: For " + "large models, collecting all sub-models can cause OOMs on the Spark driver", - Some("false"), isExpertParam = true) + Some("false"), isExpertParam = true), + ParamDesc[String]("loss", "the loss function to be optimized", finalFields = false) ) val code = genSharedParams(params) http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/main/scala/org/apache/spark/ml/param/shared/sharedParams.scala ---------------------------------------------------------------------- diff --git a/mllib/src/main/scala/org/apache/spark/ml/param/shared/sharedParams.scala b/mllib/src/main/scala/org/apache/spark/ml/param/shared/sharedParams.scala index 0004f08..13425da 100644 --- a/mllib/src/main/scala/org/apache/spark/ml/param/shared/sharedParams.scala +++ b/mllib/src/main/scala/org/apache/spark/ml/param/shared/sharedParams.scala @@ -487,4 +487,21 @@ trait HasCollectSubModels extends Params { /** @group expertGetParam */ final def getCollectSubModels: Boolean = $(collectSubModels) } + +/** + * Trait for shared param loss. This trait may be changed or + * removed between minor versions. + */ +@DeveloperApi +trait HasLoss extends Params { + + /** + * Param for the loss function to be optimized. + * @group param + */ + val loss: Param[String] = new Param[String](this, "loss", "the loss function to be optimized") + + /** @group getParam */ + final def getLoss: String = $(loss) +} // scalastyle:on http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---------------------------------------------------------------------- diff --git a/mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala b/mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala index da6bcf0..a5873d0 100644 --- a/mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala +++ b/mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala @@ -20,7 +20,7 @@ package org.apache.spark.ml.regression import scala.collection.mutable import breeze.linalg.{DenseVector => BDV} -import breeze.optimize.{CachedDiffFunction, LBFGS => BreezeLBFGS, OWLQN => BreezeOWLQN} +import breeze.optimize.{CachedDiffFunction, LBFGS => BreezeLBFGS, LBFGSB => BreezeLBFGSB, OWLQN => BreezeOWLQN} import breeze.stats.distributions.StudentsT import org.apache.hadoop.fs.Path @@ -32,9 +32,9 @@ import org.apache.spark.ml.feature.Instance import org.apache.spark.ml.linalg.{Vector, Vectors} import org.apache.spark.ml.linalg.BLAS._ import org.apache.spark.ml.optim.WeightedLeastSquares -import org.apache.spark.ml.optim.aggregator.LeastSquaresAggregator +import org.apache.spark.ml.optim.aggregator.{HuberAggregator, LeastSquaresAggregator} import org.apache.spark.ml.optim.loss.{L2Regularization, RDDLossFunction} -import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators} +import org.apache.spark.ml.param.{DoubleParam, Param, ParamMap, ParamValidators} import org.apache.spark.ml.param.shared._ import org.apache.spark.ml.util._ import org.apache.spark.mllib.evaluation.RegressionMetrics @@ -44,8 +44,9 @@ import org.apache.spark.mllib.util.MLUtils import org.apache.spark.rdd.RDD import org.apache.spark.sql.{DataFrame, Dataset, Row} import org.apache.spark.sql.functions._ -import org.apache.spark.sql.types.DoubleType +import org.apache.spark.sql.types.{DataType, DoubleType, StructType} import org.apache.spark.storage.StorageLevel +import org.apache.spark.util.VersionUtils.majorMinorVersion /** * Params for linear regression. @@ -53,7 +54,7 @@ import org.apache.spark.storage.StorageLevel private[regression] trait LinearRegressionParams extends PredictorParams with HasRegParam with HasElasticNetParam with HasMaxIter with HasTol with HasFitIntercept with HasStandardization with HasWeightCol with HasSolver - with HasAggregationDepth { + with HasAggregationDepth with HasLoss { import LinearRegression._ @@ -69,25 +70,105 @@ private[regression] trait LinearRegressionParams extends PredictorParams "The solver algorithm for optimization. Supported options: " + s"${supportedSolvers.mkString(", ")}. (Default auto)", ParamValidators.inArray[String](supportedSolvers)) + + /** + * The loss function to be optimized. + * Supported options: "squaredError" and "huber". + * Default: "squaredError" + * + * @group param + */ + @Since("2.3.0") + final override val loss: Param[String] = new Param[String](this, "loss", "The loss function to" + + s" be optimized. Supported options: ${supportedLosses.mkString(", ")}. (Default squaredError)", + ParamValidators.inArray[String](supportedLosses)) + + /** + * The shape parameter to control the amount of robustness. Must be > 1.0. + * At larger values of epsilon, the huber criterion becomes more similar to least squares + * regression; for small values of epsilon, the criterion is more similar to L1 regression. + * Default is 1.35 to get as much robustness as possible while retaining + * 95% statistical efficiency for normally distributed data. It matches sklearn + * HuberRegressor and is "M" from <a href="http://statweb.stanford.edu/~owen/reports/hhu.pdf";> + * A robust hybrid of lasso and ridge regression</a>. + * Only valid when "loss" is "huber". + * + * @group expertParam + */ + @Since("2.3.0") + final val epsilon = new DoubleParam(this, "epsilon", "The shape parameter to control the " + + "amount of robustness. Must be > 1.0.", ParamValidators.gt(1.0)) + + /** @group getExpertParam */ + @Since("2.3.0") + def getEpsilon: Double = $(epsilon) + + override protected def validateAndTransformSchema( + schema: StructType, + fitting: Boolean, + featuresDataType: DataType): StructType = { + if ($(loss) == Huber) { + require($(solver)!= Normal, "LinearRegression with huber loss doesn't support " + + "normal solver, please change solver to auto or l-bfgs.") + require($(elasticNetParam) == 0.0, "LinearRegression with huber loss only supports " + + s"L2 regularization, but got elasticNetParam = $getElasticNetParam.") + + } + super.validateAndTransformSchema(schema, fitting, featuresDataType) + } } /** * Linear regression. * - * The learning objective is to minimize the squared error, with regularization. - * The specific squared error loss function used is: - * - * <blockquote> - * $$ - * L = 1/2n ||A coefficients - y||^2^ - * $$ - * </blockquote> + * The learning objective is to minimize the specified loss function, with regularization. + * This supports two kinds of loss: + * - squaredError (a.k.a squared loss) + * - huber (a hybrid of squared error for relatively small errors and absolute error for + * relatively large ones, and we estimate the scale parameter from training data) * * This supports multiple types of regularization: * - none (a.k.a. ordinary least squares) * - L2 (ridge regression) * - L1 (Lasso) * - L2 + L1 (elastic net) + * + * The squared error objective function is: + * + * <blockquote> + * $$ + * \begin{align} + * \min_{w}\frac{1}{2n}{\sum_{i=1}^n(X_{i}w - y_{i})^{2} + + * \lambda\left[\frac{1-\alpha}{2}{||w||_{2}}^{2} + \alpha{||w||_{1}}\right]} + * \end{align} + * $$ + * </blockquote> + * + * The huber objective function is: + * + * <blockquote> + * $$ + * \begin{align} + * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma + + * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) + \frac{1}{2}\lambda {||w||_2}^2} + * \end{align} + * $$ + * </blockquote> + * + * where + * + * <blockquote> + * $$ + * \begin{align} + * H_m(z) = \begin{cases} + * z^2, & \text {if } |z| < \epsilon, \\ + * 2\epsilon|z| - \epsilon^2, & \text{otherwise} + * \end{cases} + * \end{align} + * $$ + * </blockquote> + * + * Note: Fitting with huber loss only supports none and L2 regularization. */ @Since("1.3.0") class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String) @@ -142,6 +223,9 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String * For alpha in (0,1), the penalty is a combination of L1 and L2. * Default is 0.0 which is an L2 penalty. * + * Note: Fitting with huber loss only supports None and L2 regularization, + * so throws exception if this param is non-zero value. + * * @group setParam */ @Since("1.4.0") @@ -190,6 +274,8 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String * The Normal Equations solver will be used when possible, but this will automatically fall * back to iterative optimization methods when needed. * + * Note: Fitting with huber loss doesn't support normal solver, + * so throws exception if this param was set with "normal". * @group setParam */ @Since("1.6.0") @@ -208,6 +294,26 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String def setAggregationDepth(value: Int): this.type = set(aggregationDepth, value) setDefault(aggregationDepth -> 2) + /** + * Sets the value of param [[loss]]. + * Default is "squaredError". + * + * @group setParam + */ + @Since("2.3.0") + def setLoss(value: String): this.type = set(loss, value) + setDefault(loss -> SquaredError) + + /** + * Sets the value of param [[epsilon]]. + * Default is 1.35. + * + * @group setExpertParam + */ + @Since("2.3.0") + def setEpsilon(value: Double): this.type = set(epsilon, value) + setDefault(epsilon -> 1.35) + override protected def train(dataset: Dataset[_]): LinearRegressionModel = { // Extract the number of features before deciding optimization solver. val numFeatures = dataset.select(col($(featuresCol))).first().getAs[Vector](0).size @@ -220,12 +326,12 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String } val instr = Instrumentation.create(this, dataset) - instr.logParams(labelCol, featuresCol, weightCol, predictionCol, solver, tol, - elasticNetParam, fitIntercept, maxIter, regParam, standardization, aggregationDepth) + instr.logParams(labelCol, featuresCol, weightCol, predictionCol, solver, tol, elasticNetParam, + fitIntercept, maxIter, regParam, standardization, aggregationDepth, loss, epsilon) instr.logNumFeatures(numFeatures) - if (($(solver) == Auto && - numFeatures <= WeightedLeastSquares.MAX_NUM_FEATURES) || $(solver) == Normal) { + if ($(loss) == SquaredError && (($(solver) == Auto && + numFeatures <= WeightedLeastSquares.MAX_NUM_FEATURES) || $(solver) == Normal)) { // For low dimensional data, WeightedLeastSquares is more efficient since the // training algorithm only requires one pass through the data. (SPARK-10668) @@ -330,12 +436,13 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String // Since we implicitly do the feature scaling when we compute the cost function // to improve the convergence, the effective regParam will be changed. - val effectiveRegParam = $(regParam) / yStd + val effectiveRegParam = $(loss) match { + case SquaredError => $(regParam) / yStd + case Huber => $(regParam) + } val effectiveL1RegParam = $(elasticNetParam) * effectiveRegParam val effectiveL2RegParam = (1.0 - $(elasticNetParam)) * effectiveRegParam - val getAggregatorFunc = new LeastSquaresAggregator(yStd, yMean, $(fitIntercept), - bcFeaturesStd, bcFeaturesMean)(_) val getFeaturesStd = (j: Int) => if (j >= 0 && j < numFeatures) featuresStd(j) else 0.0 val regularization = if (effectiveL2RegParam != 0.0) { val shouldApply = (idx: Int) => idx >= 0 && idx < numFeatures @@ -344,33 +451,58 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String } else { None } - val costFun = new RDDLossFunction(instances, getAggregatorFunc, regularization, - $(aggregationDepth)) - val optimizer = if ($(elasticNetParam) == 0.0 || effectiveRegParam == 0.0) { - new BreezeLBFGS[BDV[Double]]($(maxIter), 10, $(tol)) - } else { - val standardizationParam = $(standardization) - def effectiveL1RegFun = (index: Int) => { - if (standardizationParam) { - effectiveL1RegParam + val costFun = $(loss) match { + case SquaredError => + val getAggregatorFunc = new LeastSquaresAggregator(yStd, yMean, $(fitIntercept), + bcFeaturesStd, bcFeaturesMean)(_) + new RDDLossFunction(instances, getAggregatorFunc, regularization, $(aggregationDepth)) + case Huber => + val getAggregatorFunc = new HuberAggregator($(fitIntercept), $(epsilon), bcFeaturesStd)(_) + new RDDLossFunction(instances, getAggregatorFunc, regularization, $(aggregationDepth)) + } + + val optimizer = $(loss) match { + case SquaredError => + if ($(elasticNetParam) == 0.0 || effectiveRegParam == 0.0) { + new BreezeLBFGS[BDV[Double]]($(maxIter), 10, $(tol)) } else { - // If `standardization` is false, we still standardize the data - // to improve the rate of convergence; as a result, we have to - // perform this reverse standardization by penalizing each component - // differently to get effectively the same objective function when - // the training dataset is not standardized. - if (featuresStd(index) != 0.0) effectiveL1RegParam / featuresStd(index) else 0.0 + val standardizationParam = $(standardization) + def effectiveL1RegFun = (index: Int) => { + if (standardizationParam) { + effectiveL1RegParam + } else { + // If `standardization` is false, we still standardize the data + // to improve the rate of convergence; as a result, we have to + // perform this reverse standardization by penalizing each component + // differently to get effectively the same objective function when + // the training dataset is not standardized. + if (featuresStd(index) != 0.0) effectiveL1RegParam / featuresStd(index) else 0.0 + } + } + new BreezeOWLQN[Int, BDV[Double]]($(maxIter), 10, effectiveL1RegFun, $(tol)) } - } - new BreezeOWLQN[Int, BDV[Double]]($(maxIter), 10, effectiveL1RegFun, $(tol)) + case Huber => + val dim = if ($(fitIntercept)) numFeatures + 2 else numFeatures + 1 + val lowerBounds = BDV[Double](Array.fill(dim)(Double.MinValue)) + // Optimize huber loss in space "\sigma > 0" + lowerBounds(dim - 1) = Double.MinPositiveValue + val upperBounds = BDV[Double](Array.fill(dim)(Double.MaxValue)) + new BreezeLBFGSB(lowerBounds, upperBounds, $(maxIter), 10, $(tol)) + } + + val initialValues = $(loss) match { + case SquaredError => + Vectors.zeros(numFeatures) + case Huber => + val dim = if ($(fitIntercept)) numFeatures + 2 else numFeatures + 1 + Vectors.dense(Array.fill(dim)(1.0)) } - val initialCoefficients = Vectors.zeros(numFeatures) val states = optimizer.iterations(new CachedDiffFunction(costFun), - initialCoefficients.asBreeze.toDenseVector) + initialValues.asBreeze.toDenseVector) - val (coefficients, objectiveHistory) = { + val (coefficients, intercept, scale, objectiveHistory) = { /* Note that in Linear Regression, the objective history (loss + regularization) returned from optimizer is computed in the scaled space given by the following formula. @@ -396,35 +528,54 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0") override val uid: String bcFeaturesMean.destroy(blocking = false) bcFeaturesStd.destroy(blocking = false) + val parameters = state.x.toArray.clone() + /* The coefficients are trained in the scaled space; we're converting them back to the original space. */ - val rawCoefficients = state.x.toArray.clone() + val rawCoefficients: Array[Double] = $(loss) match { + case SquaredError => parameters + case Huber => parameters.slice(0, numFeatures) + } + var i = 0 val len = rawCoefficients.length + val multiplier = $(loss) match { + case SquaredError => yStd + case Huber => 1.0 + } while (i < len) { - rawCoefficients(i) *= { if (featuresStd(i) != 0.0) yStd / featuresStd(i) else 0.0 } + rawCoefficients(i) *= { if (featuresStd(i) != 0.0) multiplier / featuresStd(i) else 0.0 } i += 1 } - (Vectors.dense(rawCoefficients).compressed, arrayBuilder.result()) - } + val interceptValue: Double = if ($(fitIntercept)) { + $(loss) match { + case SquaredError => + /* + The intercept of squared error in R's GLMNET is computed using closed form + after the coefficients are converged. See the following discussion for detail. + http://stats.stackexchange.com/questions/13617/how-is-the-intercept-computed-in-glmnet + */ + yMean - dot(Vectors.dense(rawCoefficients), Vectors.dense(featuresMean)) + case Huber => parameters(numFeatures) + } + } else { + 0.0 + } - /* - The intercept in R's GLMNET is computed using closed form after the coefficients are - converged. See the following discussion for detail. - http://stats.stackexchange.com/questions/13617/how-is-the-intercept-computed-in-glmnet - */ - val intercept = if ($(fitIntercept)) { - yMean - dot(coefficients, Vectors.dense(featuresMean)) - } else { - 0.0 + val scaleValue: Double = $(loss) match { + case SquaredError => 1.0 + case Huber => parameters.last + } + + (Vectors.dense(rawCoefficients).compressed, interceptValue, scaleValue, arrayBuilder.result()) } if (handlePersistence) instances.unpersist() - val model = copyValues(new LinearRegressionModel(uid, coefficients, intercept)) + val model = copyValues(new LinearRegressionModel(uid, coefficients, intercept, scale)) // Handle possible missing or invalid prediction columns val (summaryModel, predictionColName) = model.findSummaryModelAndPredictionCol() @@ -471,6 +622,15 @@ object LinearRegression extends DefaultParamsReadable[LinearRegression] { /** Set of solvers that LinearRegression supports. */ private[regression] val supportedSolvers = Array(Auto, Normal, LBFGS) + + /** String name for "squaredError". */ + private[regression] val SquaredError = "squaredError" + + /** String name for "huber". */ + private[regression] val Huber = "huber" + + /** Set of loss function names that LinearRegression supports. */ + private[regression] val supportedLosses = Array(SquaredError, Huber) } /** @@ -480,10 +640,14 @@ object LinearRegression extends DefaultParamsReadable[LinearRegression] { class LinearRegressionModel private[ml] ( @Since("1.4.0") override val uid: String, @Since("2.0.0") val coefficients: Vector, - @Since("1.3.0") val intercept: Double) + @Since("1.3.0") val intercept: Double, + @Since("2.3.0") val scale: Double) extends RegressionModel[Vector, LinearRegressionModel] with LinearRegressionParams with MLWritable { + def this(uid: String, coefficients: Vector, intercept: Double) = + this(uid, coefficients, intercept, 1.0) + private var trainingSummary: Option[LinearRegressionTrainingSummary] = None override val numFeatures: Int = coefficients.size @@ -570,13 +734,13 @@ object LinearRegressionModel extends MLReadable[LinearRegressionModel] { private[LinearRegressionModel] class LinearRegressionModelWriter(instance: LinearRegressionModel) extends MLWriter with Logging { - private case class Data(intercept: Double, coefficients: Vector) + private case class Data(intercept: Double, coefficients: Vector, scale: Double) override protected def saveImpl(path: String): Unit = { // Save metadata and Params DefaultParamsWriter.saveMetadata(instance, path, sc) - // Save model data: intercept, coefficients - val data = Data(instance.intercept, instance.coefficients) + // Save model data: intercept, coefficients, scale + val data = Data(instance.intercept, instance.coefficients, instance.scale) val dataPath = new Path(path, "data").toString sparkSession.createDataFrame(Seq(data)).repartition(1).write.parquet(dataPath) } @@ -592,11 +756,20 @@ object LinearRegressionModel extends MLReadable[LinearRegressionModel] { val dataPath = new Path(path, "data").toString val data = sparkSession.read.format("parquet").load(dataPath) - val Row(intercept: Double, coefficients: Vector) = - MLUtils.convertVectorColumnsToML(data, "coefficients") - .select("intercept", "coefficients") - .head() - val model = new LinearRegressionModel(metadata.uid, coefficients, intercept) + val (majorVersion, minorVersion) = majorMinorVersion(metadata.sparkVersion) + val model = if (majorVersion < 2 || (majorVersion == 2 && minorVersion <= 2)) { + // Spark 2.2 and before + val Row(intercept: Double, coefficients: Vector) = + MLUtils.convertVectorColumnsToML(data, "coefficients") + .select("intercept", "coefficients") + .head() + new LinearRegressionModel(metadata.uid, coefficients, intercept) + } else { + // Spark 2.3 and later + val Row(intercept: Double, coefficients: Vector, scale: Double) = + data.select("intercept", "coefficients", "scale").head() + new LinearRegressionModel(metadata.uid, coefficients, intercept, scale) + } DefaultParamsReader.getAndSetParams(model, metadata) model http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/test/scala/org/apache/spark/ml/optim/aggregator/HuberAggregatorSuite.scala ---------------------------------------------------------------------- diff --git a/mllib/src/test/scala/org/apache/spark/ml/optim/aggregator/HuberAggregatorSuite.scala b/mllib/src/test/scala/org/apache/spark/ml/optim/aggregator/HuberAggregatorSuite.scala new file mode 100644 index 0000000..718ffa2 --- /dev/null +++ b/mllib/src/test/scala/org/apache/spark/ml/optim/aggregator/HuberAggregatorSuite.scala @@ -0,0 +1,170 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.spark.ml.optim.aggregator + +import org.apache.spark.SparkFunSuite +import org.apache.spark.ml.feature.Instance +import org.apache.spark.ml.linalg.{BLAS, Vector, Vectors} +import org.apache.spark.ml.util.TestingUtils._ +import org.apache.spark.mllib.util.MLlibTestSparkContext + +class HuberAggregatorSuite extends SparkFunSuite with MLlibTestSparkContext { + + import DifferentiableLossAggregatorSuite.getRegressionSummarizers + + @transient var instances: Array[Instance] = _ + @transient var instancesConstantFeature: Array[Instance] = _ + @transient var instancesConstantFeatureFiltered: Array[Instance] = _ + + override def beforeAll(): Unit = { + super.beforeAll() + instances = Array( + Instance(0.0, 0.1, Vectors.dense(1.0, 2.0)), + Instance(1.0, 0.5, Vectors.dense(1.5, 1.0)), + Instance(2.0, 0.3, Vectors.dense(4.0, 0.5)) + ) + instancesConstantFeature = Array( + Instance(0.0, 0.1, Vectors.dense(1.0, 2.0)), + Instance(1.0, 0.5, Vectors.dense(1.0, 1.0)), + Instance(2.0, 0.3, Vectors.dense(1.0, 0.5)) + ) + instancesConstantFeatureFiltered = Array( + Instance(0.0, 0.1, Vectors.dense(2.0)), + Instance(1.0, 0.5, Vectors.dense(1.0)), + Instance(2.0, 0.3, Vectors.dense(0.5)) + ) + } + + /** Get summary statistics for some data and create a new HuberAggregator. */ + private def getNewAggregator( + instances: Array[Instance], + parameters: Vector, + fitIntercept: Boolean, + epsilon: Double): HuberAggregator = { + val (featuresSummarizer, _) = getRegressionSummarizers(instances) + val featuresStd = featuresSummarizer.variance.toArray.map(math.sqrt) + val bcFeaturesStd = spark.sparkContext.broadcast(featuresStd) + val bcParameters = spark.sparkContext.broadcast(parameters) + new HuberAggregator(fitIntercept, epsilon, bcFeaturesStd)(bcParameters) + } + + test("aggregator add method should check input size") { + val parameters = Vectors.dense(1.0, 2.0, 3.0, 4.0) + val agg = getNewAggregator(instances, parameters, fitIntercept = true, epsilon = 1.35) + withClue("HuberAggregator features dimension must match parameters dimension") { + intercept[IllegalArgumentException] { + agg.add(Instance(1.0, 1.0, Vectors.dense(2.0))) + } + } + } + + test("negative weight") { + val parameters = Vectors.dense(1.0, 2.0, 3.0, 4.0) + val agg = getNewAggregator(instances, parameters, fitIntercept = true, epsilon = 1.35) + withClue("HuberAggregator does not support negative instance weights.") { + intercept[IllegalArgumentException] { + agg.add(Instance(1.0, -1.0, Vectors.dense(2.0, 1.0))) + } + } + } + + test("check sizes") { + val paramWithIntercept = Vectors.dense(1.0, 2.0, 3.0, 4.0) + val paramWithoutIntercept = Vectors.dense(1.0, 2.0, 4.0) + val aggIntercept = getNewAggregator(instances, paramWithIntercept, + fitIntercept = true, epsilon = 1.35) + val aggNoIntercept = getNewAggregator(instances, paramWithoutIntercept, + fitIntercept = false, epsilon = 1.35) + instances.foreach(aggIntercept.add) + instances.foreach(aggNoIntercept.add) + + assert(aggIntercept.gradient.size === 4) + assert(aggNoIntercept.gradient.size === 3) + } + + test("check correctness") { + val parameters = Vectors.dense(1.0, 2.0, 3.0, 4.0) + val numFeatures = 2 + val (featuresSummarizer, _) = getRegressionSummarizers(instances) + val featuresStd = featuresSummarizer.variance.toArray.map(math.sqrt) + val epsilon = 1.35 + val weightSum = instances.map(_.weight).sum + + val agg = getNewAggregator(instances, parameters, fitIntercept = true, epsilon) + instances.foreach(agg.add) + + // compute expected loss sum + val coefficients = parameters.toArray.slice(0, 2) + val intercept = parameters(2) + val sigma = parameters(3) + val stdCoef = coefficients.indices.map(i => coefficients(i) / featuresStd(i)).toArray + val lossSum = instances.map { case Instance(label, weight, features) => + val margin = BLAS.dot(Vectors.dense(stdCoef), features) + intercept + val linearLoss = label - margin + if (math.abs(linearLoss) <= sigma * epsilon) { + 0.5 * weight * (sigma + math.pow(linearLoss, 2.0) / sigma) + } else { + 0.5 * weight * (sigma + 2.0 * epsilon * math.abs(linearLoss) - sigma * epsilon * epsilon) + } + }.sum + val loss = lossSum / weightSum + + // compute expected gradients + val gradientCoef = new Array[Double](numFeatures + 2) + instances.foreach { case Instance(label, weight, features) => + val margin = BLAS.dot(Vectors.dense(stdCoef), features) + intercept + val linearLoss = label - margin + if (math.abs(linearLoss) <= sigma * epsilon) { + features.toArray.indices.foreach { i => + gradientCoef(i) += + -1.0 * weight * (linearLoss / sigma) * (features(i) / featuresStd(i)) + } + gradientCoef(2) += -1.0 * weight * (linearLoss / sigma) + gradientCoef(3) += 0.5 * weight * (1.0 - math.pow(linearLoss / sigma, 2.0)) + } else { + val sign = if (linearLoss >= 0) -1.0 else 1.0 + features.toArray.indices.foreach { i => + gradientCoef(i) += weight * sign * epsilon * (features(i) / featuresStd(i)) + } + gradientCoef(2) += weight * sign * epsilon + gradientCoef(3) += 0.5 * weight * (1.0 - epsilon * epsilon) + } + } + val gradient = Vectors.dense(gradientCoef.map(_ / weightSum)) + + assert(loss ~== agg.loss relTol 0.01) + assert(gradient ~== agg.gradient relTol 0.01) + } + + test("check with zero standard deviation") { + val parameters = Vectors.dense(1.0, 2.0, 3.0, 4.0) + val parametersFiltered = Vectors.dense(2.0, 3.0, 4.0) + val aggConstantFeature = getNewAggregator(instancesConstantFeature, parameters, + fitIntercept = true, epsilon = 1.35) + val aggConstantFeatureFiltered = getNewAggregator(instancesConstantFeatureFiltered, + parametersFiltered, fitIntercept = true, epsilon = 1.35) + instances.foreach(aggConstantFeature.add) + instancesConstantFeatureFiltered.foreach(aggConstantFeatureFiltered.add) + // constant features should not affect gradient + def validateGradient(grad: Vector, gradFiltered: Vector): Unit = { + assert(grad(0) === 0.0) + assert(grad(1) ~== gradFiltered(0) relTol 0.01) + } + + validateGradient(aggConstantFeature.gradient, aggConstantFeatureFiltered.gradient) + } +} http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala ---------------------------------------------------------------------- diff --git a/mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala b/mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala index aec5ac0..9bb2895 100644 --- a/mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala +++ b/mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala @@ -41,6 +41,7 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { @transient var datasetWithWeight: DataFrame = _ @transient var datasetWithWeightConstantLabel: DataFrame = _ @transient var datasetWithWeightZeroLabel: DataFrame = _ + @transient var datasetWithOutlier: DataFrame = _ override def beforeAll(): Unit = { super.beforeAll() @@ -107,6 +108,16 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { Instance(0.0, 3.0, Vectors.dense(2.0, 11.0)), Instance(0.0, 4.0, Vectors.dense(3.0, 13.0)) ), 2).toDF() + + datasetWithOutlier = { + val inlierData = LinearDataGenerator.generateLinearInput( + intercept = 6.3, weights = Array(4.7, 7.2), xMean = Array(0.9, -1.3), + xVariance = Array(0.7, 1.2), nPoints = 900, seed, eps = 0.1) + val outlierData = LinearDataGenerator.generateLinearInput( + intercept = -2.1, weights = Array(0.6, -1.2), xMean = Array(0.9, -1.3), + xVariance = Array(1.5, 0.8), nPoints = 100, seed, eps = 0.1) + sc.parallelize(inlierData ++ outlierData, 2).map(_.asML).toDF() + } } /** @@ -127,6 +138,10 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { datasetWithSparseFeature.rdd.map { case Row(label: Double, features: Vector) => label + "," + features.toArray.mkString(",") }.repartition(1).saveAsTextFile("target/tmp/LinearRegressionSuite/datasetWithSparseFeature") + + datasetWithOutlier.rdd.map { case Row(label: Double, features: Vector) => + label + "," + features.toArray.mkString(",") + }.repartition(1).saveAsTextFile("target/tmp/LinearRegressionSuite/datasetWithOutlier") } test("params") { @@ -144,7 +159,9 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { assert(lir.getElasticNetParam === 0.0) assert(lir.getFitIntercept) assert(lir.getStandardization) - assert(lir.getSolver == "auto") + assert(lir.getSolver === "auto") + assert(lir.getLoss === "squaredError") + assert(lir.getEpsilon === 1.35) val model = lir.fit(datasetWithDenseFeature) MLTestingUtils.checkCopyAndUids(lir, model) @@ -160,11 +177,27 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { assert(model.getFeaturesCol === "features") assert(model.getPredictionCol === "prediction") assert(model.intercept !== 0.0) + assert(model.scale === 1.0) assert(model.hasParent) val numFeatures = datasetWithDenseFeature.select("features").first().getAs[Vector](0).size assert(model.numFeatures === numFeatures) } + test("linear regression: illegal params") { + withClue("LinearRegression with huber loss only supports L2 regularization") { + intercept[IllegalArgumentException] { + new LinearRegression().setLoss("huber").setElasticNetParam(0.5) + .fit(datasetWithDenseFeature) + } + } + + withClue("LinearRegression with huber loss doesn't support normal solver") { + intercept[IllegalArgumentException] { + new LinearRegression().setLoss("huber").setSolver("normal").fit(datasetWithDenseFeature) + } + } + } + test("linear regression handles singular matrices") { // check for both constant columns with intercept (zero std) and collinear val singularDataConstantColumn = sc.parallelize(Seq( @@ -837,6 +870,7 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { (1.0, 0.21, true, true) ) + // For squaredError loss for (solver <- Seq("auto", "l-bfgs", "normal"); (elasticNetParam, regParam, fitIntercept, standardization) <- testParams) { val estimator = new LinearRegression() @@ -852,6 +886,22 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { MLTestingUtils.testOversamplingVsWeighting[LinearRegressionModel, LinearRegression]( datasetWithStrongNoise.as[LabeledPoint], estimator, modelEquals, seed) } + + // For huber loss + for ((_, regParam, fitIntercept, standardization) <- testParams) { + val estimator = new LinearRegression() + .setLoss("huber") + .setFitIntercept(fitIntercept) + .setStandardization(standardization) + .setRegParam(regParam) + MLTestingUtils.testArbitrarilyScaledWeights[LinearRegressionModel, LinearRegression]( + datasetWithOutlier.as[LabeledPoint], estimator, modelEquals) + MLTestingUtils.testOutliersWithSmallWeights[LinearRegressionModel, LinearRegression]( + datasetWithOutlier.as[LabeledPoint], estimator, numClasses, modelEquals, + outlierRatio = 3) + MLTestingUtils.testOversamplingVsWeighting[LinearRegressionModel, LinearRegression]( + datasetWithOutlier.as[LabeledPoint], estimator, modelEquals, seed) + } } test("linear regression model with l-bfgs with big feature datasets") { @@ -1004,6 +1054,198 @@ class LinearRegressionSuite extends MLTest with DefaultReadWriteTest { } } } + + test("linear regression (huber loss) with intercept without regularization") { + val trainer1 = (new LinearRegression).setLoss("huber") + .setFitIntercept(true).setStandardization(true) + val trainer2 = (new LinearRegression).setLoss("huber") + .setFitIntercept(true).setStandardization(false) + + val model1 = trainer1.fit(datasetWithOutlier) + val model2 = trainer2.fit(datasetWithOutlier) + + /* + Using the following Python code to load the data and train the model using + scikit-learn package. + + import pandas as pd + import numpy as np + from sklearn.linear_model import HuberRegressor + df = pd.read_csv("path", header = None) + X = df[df.columns[1:3]] + y = np.array(df[df.columns[0]]) + huber = HuberRegressor(fit_intercept=True, alpha=0.0, max_iter=100, epsilon=1.35) + huber.fit(X, y) + + >>> huber.coef_ + array([ 4.68998007, 7.19429011]) + >>> huber.intercept_ + 6.3002404351083037 + >>> huber.scale_ + 0.077810159205220747 + */ + val coefficientsPy = Vectors.dense(4.68998007, 7.19429011) + val interceptPy = 6.30024044 + val scalePy = 0.07781016 + + assert(model1.coefficients ~= coefficientsPy relTol 1E-3) + assert(model1.intercept ~== interceptPy relTol 1E-3) + assert(model1.scale ~== scalePy relTol 1E-3) + + // Without regularization, with or without standardization will converge to the same solution. + assert(model2.coefficients ~= coefficientsPy relTol 1E-3) + assert(model2.intercept ~== interceptPy relTol 1E-3) + assert(model2.scale ~== scalePy relTol 1E-3) + } + + test("linear regression (huber loss) without intercept without regularization") { + val trainer1 = (new LinearRegression).setLoss("huber") + .setFitIntercept(false).setStandardization(true) + val trainer2 = (new LinearRegression).setLoss("huber") + .setFitIntercept(false).setStandardization(false) + + val model1 = trainer1.fit(datasetWithOutlier) + val model2 = trainer2.fit(datasetWithOutlier) + + /* + huber = HuberRegressor(fit_intercept=False, alpha=0.0, max_iter=100, epsilon=1.35) + huber.fit(X, y) + + >>> huber.coef_ + array([ 6.71756703, 5.08873222]) + >>> huber.intercept_ + 0.0 + >>> huber.scale_ + 2.5560209922722317 + */ + val coefficientsPy = Vectors.dense(6.71756703, 5.08873222) + val interceptPy = 0.0 + val scalePy = 2.55602099 + + assert(model1.coefficients ~= coefficientsPy relTol 1E-3) + assert(model1.intercept === interceptPy) + assert(model1.scale ~== scalePy relTol 1E-3) + + // Without regularization, with or without standardization will converge to the same solution. + assert(model2.coefficients ~= coefficientsPy relTol 1E-3) + assert(model2.intercept === interceptPy) + assert(model2.scale ~== scalePy relTol 1E-3) + } + + test("linear regression (huber loss) with intercept with L2 regularization") { + val trainer1 = (new LinearRegression).setLoss("huber") + .setFitIntercept(true).setRegParam(0.21).setStandardization(true) + val trainer2 = (new LinearRegression).setLoss("huber") + .setFitIntercept(true).setRegParam(0.21).setStandardization(false) + + val model1 = trainer1.fit(datasetWithOutlier) + val model2 = trainer2.fit(datasetWithOutlier) + + /* + Since scikit-learn HuberRegressor does not support standardization, + we do it manually out of the estimator. + + xStd = np.std(X, axis=0) + scaledX = X / xStd + huber = HuberRegressor(fit_intercept=True, alpha=210, max_iter=100, epsilon=1.35) + huber.fit(scaledX, y) + + >>> np.array(huber.coef_ / xStd) + array([ 1.97732633, 3.38816722]) + >>> huber.intercept_ + 3.7527581430531227 + >>> huber.scale_ + 3.787363673371801 + */ + val coefficientsPy1 = Vectors.dense(1.97732633, 3.38816722) + val interceptPy1 = 3.75275814 + val scalePy1 = 3.78736367 + + assert(model1.coefficients ~= coefficientsPy1 relTol 1E-2) + assert(model1.intercept ~== interceptPy1 relTol 1E-2) + assert(model1.scale ~== scalePy1 relTol 1E-2) + + /* + huber = HuberRegressor(fit_intercept=True, alpha=210, max_iter=100, epsilon=1.35) + huber.fit(X, y) + + >>> huber.coef_ + array([ 1.73346444, 3.63746999]) + >>> huber.intercept_ + 4.3017134790781739 + >>> huber.scale_ + 3.6472742809286793 + */ + val coefficientsPy2 = Vectors.dense(1.73346444, 3.63746999) + val interceptPy2 = 4.30171347 + val scalePy2 = 3.64727428 + + assert(model2.coefficients ~= coefficientsPy2 relTol 1E-3) + assert(model2.intercept ~== interceptPy2 relTol 1E-3) + assert(model2.scale ~== scalePy2 relTol 1E-3) + } + + test("linear regression (huber loss) without intercept with L2 regularization") { + val trainer1 = (new LinearRegression).setLoss("huber") + .setFitIntercept(false).setRegParam(0.21).setStandardization(true) + val trainer2 = (new LinearRegression).setLoss("huber") + .setFitIntercept(false).setRegParam(0.21).setStandardization(false) + + val model1 = trainer1.fit(datasetWithOutlier) + val model2 = trainer2.fit(datasetWithOutlier) + + /* + Since scikit-learn HuberRegressor does not support standardization, + we do it manually out of the estimator. + + xStd = np.std(X, axis=0) + scaledX = X / xStd + huber = HuberRegressor(fit_intercept=False, alpha=210, max_iter=100, epsilon=1.35) + huber.fit(scaledX, y) + + >>> np.array(huber.coef_ / xStd) + array([ 2.59679008, 2.26973102]) + >>> huber.intercept_ + 0.0 + >>> huber.scale_ + 4.5766311924091791 + */ + val coefficientsPy1 = Vectors.dense(2.59679008, 2.26973102) + val interceptPy1 = 0.0 + val scalePy1 = 4.57663119 + + assert(model1.coefficients ~= coefficientsPy1 relTol 1E-2) + assert(model1.intercept === interceptPy1) + assert(model1.scale ~== scalePy1 relTol 1E-2) + + /* + huber = HuberRegressor(fit_intercept=False, alpha=210, max_iter=100, epsilon=1.35) + huber.fit(X, y) + + >>> huber.coef_ + array([ 2.28423908, 2.25196887]) + >>> huber.intercept_ + 0.0 + >>> huber.scale_ + 4.5979643506051753 + */ + val coefficientsPy2 = Vectors.dense(2.28423908, 2.25196887) + val interceptPy2 = 0.0 + val scalePy2 = 4.59796435 + + assert(model2.coefficients ~= coefficientsPy2 relTol 1E-3) + assert(model2.intercept === interceptPy2) + assert(model2.scale ~== scalePy2 relTol 1E-3) + } + + test("huber loss model match squared error for large epsilon") { + val trainer1 = new LinearRegression().setLoss("huber").setEpsilon(1E5) + val model1 = trainer1.fit(datasetWithOutlier) + val trainer2 = new LinearRegression() + val model2 = trainer2.fit(datasetWithOutlier) + assert(model1.coefficients ~== model2.coefficients relTol 1E-3) + assert(model1.intercept ~== model2.intercept relTol 1E-3) + } } object LinearRegressionSuite { http://git-wip-us.apache.org/repos/asf/spark/blob/1e44dd00/project/MimaExcludes.scala ---------------------------------------------------------------------- diff --git a/project/MimaExcludes.scala b/project/MimaExcludes.scala index 9be01f6..9902fed 100644 --- a/project/MimaExcludes.scala +++ b/project/MimaExcludes.scala @@ -1065,6 +1065,11 @@ object MimaExcludes { // [SPARK-21680][ML][MLLIB]optimzie Vector coompress ProblemFilters.exclude[ReversedMissingMethodProblem]("org.apache.spark.mllib.linalg.Vector.toSparseWithSize"), ProblemFilters.exclude[ReversedMissingMethodProblem]("org.apache.spark.ml.linalg.Vector.toSparseWithSize") + ) ++ Seq( + // [SPARK-3181][ML]Implement huber loss for LinearRegression. + ProblemFilters.exclude[InheritedNewAbstractMethodProblem]("org.apache.spark.ml.param.shared.HasLoss.org$apache$spark$ml$param$shared$HasLoss$_setter_$loss_="), + ProblemFilters.exclude[InheritedNewAbstractMethodProblem]("org.apache.spark.ml.param.shared.HasLoss.getLoss"), + ProblemFilters.exclude[InheritedNewAbstractMethodProblem]("org.apache.spark.ml.param.shared.HasLoss.loss") ) } --------------------------------------------------------------------- To unsubscribe, e-mail: commits-unsubscr...@spark.apache.org For additional commands, e-mail: commits-h...@spark.apache.org
