[GitHub] spark issue #12066: [SPARK-7424] [ML] ML ClassificationModel should add meta...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/12066 @holdenk Sorry for late response, I'm really busy recently. Sure, I'll close this now. Feel free to take over it. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #12066: [SPARK-7424] [ML] ML ClassificationModel should a...
Github user yanboliang closed the pull request at: https://github.com/apache/spark/pull/12066 --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #21494: [WIP][SPARK-24375][Prototype] Support barrier sch...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/21494#discussion_r194172809
--- Diff:
core/src/main/scala/org/apache/spark/scheduler/TaskSetManager.scala ---
@@ -123,6 +124,21 @@ private[spark] class TaskSetManager(
// TODO: We should kill any running task attempts when the task set
manager becomes a zombie.
private[scheduler] var isZombie = false
+ private[scheduler] lazy val barrierCoordinator = {
--- End diff --
+1 @galv
We also have ```barrierCoordinator``` with type ```RpcEndpointRef``` at
each TaskContext, so it's better to add return type for both.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #21494: [WIP][SPARK-24375][Prototype] Support barrier sch...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/21494#discussion_r194176991
--- Diff:
core/src/main/scala/org/apache/spark/barrier/BarrierCoordinator.scala ---
@@ -0,0 +1,78 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+
+package org.apache.spark.barrier
+
+import java.util.{Timer, TimerTask}
+
+import org.apache.spark.rpc.{RpcCallContext, RpcEnv, ThreadSafeRpcEndpoint}
+
+class BarrierCoordinator(
+numTasks: Int,
+timeout: Long,
+override val rpcEnv: RpcEnv) extends ThreadSafeRpcEndpoint {
+
+ private var epoch = 0
--- End diff --
Will ```epoch``` value be logged on driver and executors? It should be
useful to diagnose upper level MPI program.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #21249: [SPARK-23291][R][FOLLOWUP] Update SparkR migration note ...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/21249 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #20459: [SPARK-23107][ML] ML 2.3 QA: New Scala APIs, docs...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/20459#discussion_r165239367
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/feature/QuantileDiscretizer.scala ---
@@ -93,7 +93,7 @@ private[feature] trait QuantileDiscretizerBase extends
Params
* @group param
*/
@Since("2.1.0")
- override val handleInvalid: Param[String] = new Param[String](this,
"handleInvalid",
+ final override val handleInvalid: Param[String] = new
Param[String](this, "handleInvalid",
--- End diff --
Fair enough. I will leave ```handleInvalid``` in all estimators
```non-final```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #20459: [SPARK-23107][ML] ML 2.3 QA: New Scala APIs, docs...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/20459#discussion_r165189663
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/feature/QuantileDiscretizer.scala ---
@@ -93,7 +93,7 @@ private[feature] trait QuantileDiscretizerBase extends
Params
* @group param
*/
@Since("2.1.0")
- override val handleInvalid: Param[String] = new Param[String](this,
"handleInvalid",
+ final override val handleInvalid: Param[String] = new
Param[String](this, "handleInvalid",
--- End diff --
For this and the followings who added before 2.2, this involves breaking
change in a way, but I think we should keep them ```final``` to prevent being
changed(as we did for other param variables). cc @MLnick @WeichenXu123
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #20459: [SPARK-23107][ML] ML 2.3 QA: New Scala APIs, docs...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/20459 [SPARK-23107][ML] ML 2.3 QA: New Scala APIs, docs. ## What changes were proposed in this pull request? Audit new APIs and docs in 2.3.0. ## How was this patch tested? No test. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark SPARK-23107 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/20459.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #20459 commit c24cc140563e5807e0a1fa6754e7ae461c4d7e23 Author: Yanbo Liang <ybliang8@...> Date: 2018-01-31T20:51:34Z ML 2.3 QA: New Scala APIs, docs. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interface of d...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19156 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19994: [SPARK-22810][ML][PySpark] Expose Python API for LinearR...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19994 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r158138431
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/stat/SummarizerSuite.scala ---
@@ -35,237 +34,252 @@ class SummarizerSuite extends SparkFunSuite with
MLlibTestSparkContext {
import SummaryBuilderImpl._
private case class ExpectedMetrics(
- mean: Seq[Double],
- variance: Seq[Double],
+ mean: Vector,
+ variance: Vector,
count: Long,
- numNonZeros: Seq[Long],
- max: Seq[Double],
- min: Seq[Double],
- normL2: Seq[Double],
- normL1: Seq[Double])
+ numNonZeros: Vector,
+ max: Vector,
+ min: Vector,
+ normL2: Vector,
+ normL1: Vector)
/**
- * The input is expected to be either a sparse vector, a dense vector or
an array of doubles
- * (which will be converted to a dense vector)
- * The expected is the list of all the known metrics.
+ * The input is expected to be either a sparse vector, a dense vector.
*
- * The tests take an list of input vectors and a list of all the summary
values that
- * are expected for this input. They currently test against some fixed
subset of the
- * metrics, but should be made fuzzy in the future.
+ * The tests take an list of input vectors, and compare results with
+ * `mllib.stat.MultivariateOnlineSummarizer`. They currently test
against some fixed subset
+ * of the metrics, but should be made fuzzy in the future.
*/
- private def testExample(name: String, input: Seq[Any], exp:
ExpectedMetrics): Unit = {
+ private def testExample(name: String, inputVec: Seq[(Vector, Double)],
+ exp: ExpectedMetrics, expWithoutWeight: ExpectedMetrics): Unit = {
-def inputVec: Seq[Vector] = input.map {
- case x: Array[Double @unchecked] => Vectors.dense(x)
- case x: Seq[Double @unchecked] => Vectors.dense(x.toArray)
- case x: Vector => x
- case x => throw new Exception(x.toString)
+val summarizer = {
+ val _summarizer = new MultivariateOnlineSummarizer
+ inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v._1), v._2))
+ _summarizer
}
-val summarizer = {
+val summarizerWithoutWeight = {
val _summarizer = new MultivariateOnlineSummarizer
- inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v)))
+ inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v._1)))
_summarizer
}
// Because the Spark context is reset between tests, we cannot hold a
reference onto it.
def wrappedInit() = {
- val df = inputVec.map(Tuple1.apply).toDF("features")
- val col = df.col("features")
- (df, col)
+ val df = inputVec.toDF("features", "weight")
+ val featuresCol = df.col("features")
+ val weightCol = df.col("weight")
+ (df, featuresCol, weightCol)
}
registerTest(s"$name - mean only") {
- val (df, c) = wrappedInit()
- compare(df.select(metrics("mean").summary(c), mean(c)),
Seq(Row(exp.mean), summarizer.mean))
+ val (df, c, w) = wrappedInit()
+ compareRow(df.select(metrics("mean").summary(c, w), mean(c,
w)).first(),
+Row(Row(summarizer.mean), exp.mean))
}
-registerTest(s"$name - mean only (direct)") {
- val (df, c) = wrappedInit()
- compare(df.select(mean(c)), Seq(exp.mean))
+registerTest(s"$name - mean only w/o weight") {
+ val (df, c, _) = wrappedInit()
+ compareRow(df.select(metrics("mean").summary(c), mean(c)).first(),
+Row(Row(summarizerWithoutWeight.mean), expWithoutWeight.mean))
}
registerTest(s"$name - variance only") {
- val (df, c) = wrappedInit()
- compare(df.select(metrics("variance").summary(c), variance(c)),
-Seq(Row(exp.variance), summarizer.variance))
+ val (df, c, w) = wrappedInit()
+ compareRow(df.select(metrics("variance").summary(c, w), variance(c,
w)).first(),
+Row(Row(summarizer.variance), exp.variance))
}
-registerTest(s"$name - variance only (direct)") {
- val (df, c) = wrappedInit()
- compare(df.select(variance(c)), Seq(summarizer.variance))
+registerTest(s"$name - variance only w/o weight") {
+ val (df, c, _) = wrappedInit()
+ compareRow(df.select(metrics("variance").summary(c),
variance(c)).first(),
+Row(Row(summarize
[GitHub] spark issue #17146: [SPARK-19806][ML][PySpark] PySpark GeneralizedLinearRegr...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/17146 @Antoinelypro Sorry for late response. Actually we have default value if users don't set _link_ explicitly. Could you show the detail of your error case? Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19994: [SPARK-22810][ML][PySpark] Expose Python API for ...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/19994 [SPARK-22810][ML][PySpark] Expose Python API for LinearRegression with huber loss. ## What changes were proposed in this pull request? Expose Python API for _LinearRegression_ with _huber_ loss. ## How was this patch tested? Unit test. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark spark-22810 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19994.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19994 commit 1ed46a2ea0fe28e173df4bc9bfec301beafc1acd Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-12-15T19:58:55Z Expose Python API for LinearRegression with huber loss. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 Merged into master, thanks for all your reviewing. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r156856635
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -480,10 +640,14 @@ object LinearRegression extends
DefaultParamsReadable[LinearRegression] {
class LinearRegressionModel private[ml] (
@Since("1.4.0") override val uid: String,
@Since("2.0.0") val coefficients: Vector,
-@Since("1.3.0") val intercept: Double)
+@Since("1.3.0") val intercept: Double,
+@Since("2.3.0") val scale: Double)
extends RegressionModel[Vector, LinearRegressionModel]
with LinearRegressionParams with MLWritable {
+ def this(uid: String, coefficients: Vector, intercept: Double) =
+this(uid, coefficients, intercept, 1.0)
--- End diff --
```scale``` denotes that ```|y - X'w - c|``` is scaled down, I think it
make sense to be set 1.0 for least squares regression.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r156564056
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/stat/SummarizerSuite.scala ---
@@ -205,67 +207,21 @@ class SummarizerSuite extends SparkFunSuite with
MLlibTestSparkContext {
}
}
- test("debugging test") {
-val df = denseData(Nil)
-val c = df.col("features")
-val c1 = metrics("mean").summary(c)
-val res = df.select(c1)
-intercept[SparkException] {
- compare(res, Seq.empty)
-}
- }
-
- test("basic error handling") {
-val df = denseData(Nil)
-val c = df.col("features")
-val res = df.select(metrics("mean").summary(c), mean(c))
-intercept[SparkException] {
- compare(res, Seq.empty)
-}
- }
+ testExample("single element", Seq((Vectors.dense(0.0, 1.0, 2.0), 2.0)))
- test("no element, working metrics") {
-val df = denseData(Nil)
-val c = df.col("features")
-val res = df.select(metrics("count").summary(c), count(c))
-compare(res, Seq(Row(0L), 0L))
- }
+ testExample("multiple elements (dense)",
+Seq(
+ (Vectors.dense(-1.0, 0.0, 6.0), 0.5),
+ (Vectors.dense(3.0, -3.0, 0.0), 2.8),
+ (Vectors.dense(1.0, -3.0, 0.0), 0.0)
+)
+ )
- val singleElem = Seq(0.0, 1.0, 2.0)
- testExample("single element", Seq(singleElem), ExpectedMetrics(
-mean = singleElem,
-variance = Seq(0.0, 0.0, 0.0),
-count = 1,
-numNonZeros = Seq(0, 1, 1),
-max = singleElem,
-min = singleElem,
-normL1 = singleElem,
-normL2 = singleElem
- ))
-
- testExample("two elements", Seq(Seq(0.0, 1.0, 2.0), Seq(0.0, -1.0,
-2.0)), ExpectedMetrics(
-mean = Seq(0.0, 0.0, 0.0),
-// TODO: I have a doubt about these values, they are not normalized.
-variance = Seq(0.0, 2.0, 8.0),
-count = 2,
-numNonZeros = Seq(0, 2, 2),
-max = Seq(0.0, 1.0, 2.0),
-min = Seq(0.0, -1.0, -2.0),
-normL1 = Seq(0.0, 2.0, 4.0),
-normL2 = Seq(0.0, math.sqrt(2.0), math.sqrt(2.0) * 2.0)
- ))
-
- testExample("dense vector input",
-Seq(Seq(-1.0, 0.0, 6.0), Seq(3.0, -3.0, 0.0)),
--- End diff --
Why do you remove the test against ground true value?
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r156564200
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/stat/SummarizerSuite.scala ---
@@ -19,149 +19,165 @@ package org.apache.spark.ml.stat
import org.scalatest.exceptions.TestFailedException
-import org.apache.spark.{SparkException, SparkFunSuite}
+import org.apache.spark.SparkFunSuite
import org.apache.spark.ml.linalg.{Vector, Vectors}
import org.apache.spark.ml.util.TestingUtils._
import org.apache.spark.mllib.linalg.{Vector => OldVector, Vectors =>
OldVectors}
import org.apache.spark.mllib.stat.{MultivariateOnlineSummarizer,
Statistics}
import org.apache.spark.mllib.util.MLlibTestSparkContext
import org.apache.spark.sql.{DataFrame, Row}
-import org.apache.spark.sql.catalyst.expressions.GenericRowWithSchema
class SummarizerSuite extends SparkFunSuite with MLlibTestSparkContext {
import testImplicits._
import Summarizer._
import SummaryBuilderImpl._
- private case class ExpectedMetrics(
- mean: Seq[Double],
- variance: Seq[Double],
- count: Long,
- numNonZeros: Seq[Long],
- max: Seq[Double],
- min: Seq[Double],
- normL2: Seq[Double],
- normL1: Seq[Double])
-
/**
- * The input is expected to be either a sparse vector, a dense vector or
an array of doubles
- * (which will be converted to a dense vector)
- * The expected is the list of all the known metrics.
+ * The input is expected to be either a sparse vector, a dense vector.
*
- * The tests take an list of input vectors and a list of all the summary
values that
- * are expected for this input. They currently test against some fixed
subset of the
- * metrics, but should be made fuzzy in the future.
+ * The tests take an list of input vectors, and compare results with
+ * `mllib.stat.MultivariateOnlineSummarizer`. They currently test
against some fixed subset
+ * of the metrics, but should be made fuzzy in the future.
*/
- private def testExample(name: String, input: Seq[Any], exp:
ExpectedMetrics): Unit = {
+ private def testExample(name: String, inputVec: Seq[(Vector, Double)]):
Unit = {
-def inputVec: Seq[Vector] = input.map {
- case x: Array[Double @unchecked] => Vectors.dense(x)
- case x: Seq[Double @unchecked] => Vectors.dense(x.toArray)
- case x: Vector => x
- case x => throw new Exception(x.toString)
+val summarizer = {
+ val _summarizer = new MultivariateOnlineSummarizer
+ inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v._1), v._2))
+ _summarizer
}
-val summarizer = {
+val summarizerWithoutWeight = {
val _summarizer = new MultivariateOnlineSummarizer
- inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v)))
+ inputVec.foreach(v => _summarizer.add(OldVectors.fromML(v._1)))
_summarizer
}
// Because the Spark context is reset between tests, we cannot hold a
reference onto it.
def wrappedInit() = {
- val df = inputVec.map(Tuple1.apply).toDF("features")
- val col = df.col("features")
- (df, col)
+ val df = inputVec.toDF("features", "weight")
+ val featuresCol = df.col("features")
+ val weightCol = df.col("weight")
+ (df, featuresCol, weightCol)
}
registerTest(s"$name - mean only") {
- val (df, c) = wrappedInit()
- compare(df.select(metrics("mean").summary(c), mean(c)),
Seq(Row(exp.mean), summarizer.mean))
+ val (df, c, weight) = wrappedInit()
+ compare(df.select(metrics("mean").summary(c, weight), mean(c,
weight)),
+Seq(Row(summarizer.mean), summarizer.mean))
}
-registerTest(s"$name - mean only (direct)") {
- val (df, c) = wrappedInit()
- compare(df.select(mean(c)), Seq(exp.mean))
+registerTest(s"$name - mean only w/o weight") {
+ val (df, c, _) = wrappedInit()
+ compare(df.select(metrics("mean").summary(c), mean(c)),
+Seq(Row(summarizerWithoutWeight.mean),
summarizerWithoutWeight.mean))
}
registerTest(s"$name - variance only") {
- val (df, c) = wrappedInit()
- compare(df.select(metrics("variance").summary(c), variance(c)),
-Seq(Row(exp.variance), summarizer.variance))
+ val (df, c, weight) = wrappedInit()
--- End diff --
nit: ```weight``` can be abbreviated to ```w```.
[GitHub] spark issue #19958: [SPARK-21087] [ML] [FOLLOWUP] Sync SharedParamsCodeGen a...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19958 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19958: [SPARK-21087] [ML] [FOLLOWUP] Sync SharedParamsCodeGen a...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19958 cc @WeichenXu123 @jkbradley --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19958: [SPARK-21087] [ML] [FOLLOWUP] Sync SharedParamsCo...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/19958 [SPARK-21087] [ML] [FOLLOWUP] Sync SharedParamsCodeGen and sharedParams. ## What changes were proposed in this pull request? #19208 modified ```sharedParams.scala```, but didn't generated by ```SharedParamsCodeGen.scala```. This will involves mismatch between them. ## How was this patch tested? Existing test. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark spark-21087 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19958.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19958 commit d677ab1792542590b5317cb7c66ab56a2bde Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-12-12T22:56:38Z Sync SharedParamsCodeGen and sharedParams. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r156517313
--- Diff: mllib/src/main/scala/org/apache/spark/ml/stat/Summarizer.scala ---
@@ -197,14 +240,14 @@ private[ml] object SummaryBuilderImpl extends Logging
{
* metrics that need to de computed internally to get the final result.
*/
private val allMetrics: Seq[(String, Metric, DataType,
Seq[ComputeMetric])] = Seq(
-("mean", Mean, arrayDType, Seq(ComputeMean, ComputeWeightSum)),
-("variance", Variance, arrayDType, Seq(ComputeWeightSum, ComputeMean,
ComputeM2n)),
+("mean", Mean, vectorUDT, Seq(ComputeMean, ComputeWeightSum)),
+("variance", Variance, vectorUDT, Seq(ComputeWeightSum, ComputeMean,
ComputeM2n)),
("count", Count, LongType, Seq()),
-("numNonZeros", NumNonZeros, arrayLType, Seq(ComputeNNZ)),
-("max", Max, arrayDType, Seq(ComputeMax, ComputeNNZ)),
-("min", Min, arrayDType, Seq(ComputeMin, ComputeNNZ)),
-("normL2", NormL2, arrayDType, Seq(ComputeM2)),
-("normL1", NormL1, arrayDType, Seq(ComputeL1))
+("numNonZeros", NumNonZeros, vectorUDT, Seq(ComputeNNZ)),
--- End diff --
In the old ```mllib.stat.MultivariateOnlineSummarizer```, the internal
variable is type of ```Array[Long]```, but the return type is ```Vector```. Do
you know the impact of using ```Vector``` internal? Thanks.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19793: [SPARK-22574] [Mesos] [Submit] Check submission request ...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19793 @vanzin It seems this PR breaks [Jenkins test](https://amplab.cs.berkeley.edu/jenkins/job/SparkPullRequestBuilder/84790/console), could you help to resolve it? Also cc @gatorsmile @cloud-fan. Thanks. ``` Running Scala style checks Scalastyle checks failed at following occurrences: [error] /home/jenkins/workspace/SparkPullRequestBuilder@2/resource-managers/mesos/src/main/scala/org/apache/spark/deploy/rest/mesos/MesosRestServer.scala:89: File line length exceeds 100 characters [error] Total time: 17 s, completed Dec 12, 2017 1:29:36 PM [error] running /home/jenkins/workspace/SparkPullRequestBuilder@2/dev/lint-scala ; received return code 1 ``` --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19525: [SPARK-22289] [ML] Add JSON support for Matrix parameter...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19525 Merged into master and branch-2.2. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19676: [SPARK-14516][FOLLOWUP] Adding ClusteringEvaluato...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19676#discussion_r155913190
--- Diff:
examples/src/main/java/org/apache/spark/examples/ml/JavaKMeansExample.java ---
@@ -51,9 +52,17 @@ public static void main(String[] args) {
KMeans kmeans = new KMeans().setK(2).setSeed(1L);
KMeansModel model = kmeans.fit(dataset);
-// Evaluate clustering by computing Within Set Sum of Squared Errors.
-double WSSSE = model.computeCost(dataset);
-System.out.println("Within Set Sum of Squared Errors = " + WSSSE);
+// Make predictions
+Dataset predictions = model.transform(dataset);
+
+// Evaluate clustering by computing Silhouette score
+ClusteringEvaluator evaluator = new ClusteringEvaluator()
+ .setFeaturesCol("features")
+ .setPredictionCol("prediction")
--- End diff --
We use default values here, so it's not necessary to set them explicitly.
We should keep examples as simple as possible. Thanks.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19676: [SPARK-14516][FOLLOWUP] Adding ClusteringEvaluator to ex...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19676 It's good to have this, sorry for late response, I will make a pass tomorrow. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r155413347
--- Diff: mllib/src/main/scala/org/apache/spark/ml/param/params.scala ---
@@ -122,17 +124,33 @@ private[ml] object Param {
/** Decodes a param value from JSON. */
def jsonDecode[T](json: String): T = {
-parse(json) match {
+val jValue = parse(json)
+jValue match {
case JString(x) =>
x.asInstanceOf[T]
case JObject(v) =>
val keys = v.map(_._1)
-assert(keys.contains("type") && keys.contains("values"),
- s"Expect a JSON serialized vector but cannot find fields 'type'
and 'values' in $json.")
-JsonVectorConverter.fromJson(json).asInstanceOf[T]
+if (keys.contains("class")) {
+ implicit val formats = DefaultFormats
+ val className = (jValue \ "class").extract[String]
+ className match {
+case JsonMatrixConverter.className =>
+ val checkFields = Array("numRows", "numCols", "values",
"isTransposed")
+ require(checkFields.forall(keys.contains), s"Expect a JSON
serialized Matrix" +
+s" but cannot find fields ${checkFields.mkString(", ")} in
$json.")
+ JsonMatrixConverter.fromJson(json).asInstanceOf[T]
+
+case s => throw new SparkException(s"unrecognized class $s in
$json")
+ }
+} else { // Vector does not have class info in json
--- End diff --
I'd suggest to add more comment here to clarify why vector doesn't have
_class_ info in json, it should facilitate the code maintenance.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r155411609
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/linalg/JsonMatrixConverter.scala ---
@@ -0,0 +1,79 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.linalg
+
+import org.json4s.DefaultFormats
+import org.json4s.JsonDSL._
+import org.json4s.jackson.JsonMethods.{compact, parse => parseJson, render}
+
+private[ml] object JsonMatrixConverter {
+
+ /** Unique class name for identifying JSON object encoded by this class.
*/
+ val className = "org.apache.spark.ml.linalg.Matrix"
--- End diff --
@hhbyyh You have got a point there, agree.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r155412287
--- Diff: mllib/src/main/scala/org/apache/spark/ml/param/params.scala ---
@@ -122,17 +124,33 @@ private[ml] object Param {
/** Decodes a param value from JSON. */
def jsonDecode[T](json: String): T = {
-parse(json) match {
+val jValue = parse(json)
+jValue match {
case JString(x) =>
x.asInstanceOf[T]
case JObject(v) =>
val keys = v.map(_._1)
-assert(keys.contains("type") && keys.contains("values"),
- s"Expect a JSON serialized vector but cannot find fields 'type'
and 'values' in $json.")
-JsonVectorConverter.fromJson(json).asInstanceOf[T]
+if (keys.contains("class")) {
+ implicit val formats = DefaultFormats
+ val className = (jValue \ "class").extract[String]
+ className match {
+case JsonMatrixConverter.className =>
+ val checkFields = Array("numRows", "numCols", "values",
"isTransposed")
--- End diff --
Should we check _type_ as well?
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r155414284
--- Diff:
mllib-local/src/main/scala/org/apache/spark/ml/linalg/Matrices.scala ---
@@ -476,6 +476,10 @@ class DenseMatrix @Since("2.0.0") (
@Since("2.0.0")
object DenseMatrix {
+ @Since("2.3.0")
+ private[ml] def unapply(dm: DenseMatrix): Option[(Int, Int,
Array[Double], Boolean)] =
--- End diff --
I'm neutral on this issue. It's ok to let it private but should remove
```Since```. We can make it public later when there is clear requirement.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r155414954
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/classification/LogisticRegressionSuite.scala
---
@@ -2769,6 +2769,20 @@ class LogisticRegressionSuite
LogisticRegressionSuite.allParamSettings, checkModelData)
}
+ test("read/write with BoundsOnCoefficients") {
+def checkModelData(model: LogisticRegressionModel, model2:
LogisticRegressionModel): Unit = {
+ assert(model.getLowerBoundsOnCoefficients ===
model2.getLowerBoundsOnCoefficients)
+ assert(model.getUpperBoundsOnCoefficients ===
model2.getUpperBoundsOnCoefficients)
--- End diff --
Sure, you can update existing read/write test or your test, as long as to
check model itself rather than parameters.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r149823481
--- Diff: mllib/src/main/scala/org/apache/spark/ml/stat/Summarizer.scala ---
@@ -197,14 +240,14 @@ private[ml] object SummaryBuilderImpl extends Logging
{
* metrics that need to de computed internally to get the final result.
*/
private val allMetrics: Seq[(String, Metric, DataType,
Seq[ComputeMetric])] = Seq(
-("mean", Mean, arrayDType, Seq(ComputeMean, ComputeWeightSum)),
-("variance", Variance, arrayDType, Seq(ComputeWeightSum, ComputeMean,
ComputeM2n)),
+("mean", Mean, vectorUDT, Seq(ComputeMean, ComputeWeightSum)),
+("variance", Variance, vectorUDT, Seq(ComputeWeightSum, ComputeMean,
ComputeM2n)),
("count", Count, LongType, Seq()),
-("numNonZeros", NumNonZeros, arrayLType, Seq(ComputeNNZ)),
-("max", Max, arrayDType, Seq(ComputeMax, ComputeNNZ)),
-("min", Min, arrayDType, Seq(ComputeMin, ComputeNNZ)),
-("normL2", NormL2, arrayDType, Seq(ComputeM2)),
-("normL1", NormL1, arrayDType, Seq(ComputeL1))
+("numNonZeros", NumNonZeros, vectorUDT, Seq(ComputeNNZ)),
--- End diff --
Could you let me know why did you make this change? I think we should use
long array rather than double array to store ```numNonZeros```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interfa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19156#discussion_r149764998
--- Diff: mllib/src/main/scala/org/apache/spark/ml/stat/Summarizer.scala ---
@@ -94,46 +97,86 @@ object Summarizer extends Logging {
* - min: the minimum for each coefficient.
* - normL2: the Euclidian norm for each coefficient.
* - normL1: the L1 norm of each coefficient (sum of the absolute
values).
- * @param firstMetric the metric being provided
- * @param metrics additional metrics that can be provided.
+ * @param metrics metrics that can be provided.
* @return a builder.
* @throws IllegalArgumentException if one of the metric names is not
understood.
*
* Note: Currently, the performance of this interface is about 2x~3x
slower then using the RDD
* interface.
*/
@Since("2.3.0")
- def metrics(firstMetric: String, metrics: String*): SummaryBuilder = {
-val (typedMetrics, computeMetrics) =
getRelevantMetrics(Seq(firstMetric) ++ metrics)
+ def metrics(metrics: String*): SummaryBuilder = {
--- End diff --
+1 @cloud-fan
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19648: [SPARK-14516][ML][FOLLOW-UP] Move ClusteringEvaluatorSui...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19648 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149522834
--- Diff:
mllib-local/src/main/scala/org/apache/spark/ml/linalg/Matrices.scala ---
@@ -827,6 +831,11 @@ class SparseMatrix @Since("2.0.0") (
@Since("2.0.0")
object SparseMatrix {
+ @Since("2.3.0")
+ private[ml] def unapply(
--- End diff --
Ditto
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149533876
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/classification/LogisticRegressionSuite.scala
---
@@ -2769,6 +2769,20 @@ class LogisticRegressionSuite
LogisticRegressionSuite.allParamSettings, checkModelData)
}
+ test("read/write with BoundsOnCoefficients") {
+def checkModelData(model: LogisticRegressionModel, model2:
LogisticRegressionModel): Unit = {
+ assert(model.getLowerBoundsOnCoefficients ===
model2.getLowerBoundsOnCoefficients)
+ assert(model.getUpperBoundsOnCoefficients ===
model2.getUpperBoundsOnCoefficients)
--- End diff --
In ```checkModelData```, we should check model itself like
```coefficients``` and ```intercept```. We already have check equality for all
params including ```lowerBoundsOnCoefficients``` inside of
```testEstimatorAndModelReadWrite```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149522660
--- Diff:
mllib-local/src/main/scala/org/apache/spark/ml/linalg/Matrices.scala ---
@@ -476,6 +476,10 @@ class DenseMatrix @Since("2.0.0") (
@Since("2.0.0")
object DenseMatrix {
+ @Since("2.3.0")
+ private[ml] def unapply(dm: DenseMatrix): Option[(Int, Int,
Array[Double], Boolean)] =
--- End diff --
If this is a public API, we should remove ```private[ml]```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149530436
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/linalg/JsonMatrixConverter.scala ---
@@ -0,0 +1,79 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.linalg
+
+import org.json4s.DefaultFormats
+import org.json4s.JsonDSL._
+import org.json4s.jackson.JsonMethods.{compact, parse => parseJson, render}
+
+private[ml] object JsonMatrixConverter {
+
+ /** Unique class name for identifying JSON object encoded by this class.
*/
+ val className = "org.apache.spark.ml.linalg.Matrix"
--- End diff --
I'd suggest a more shorter string(or integer) to identify this is a matrix,
it should be huge burden to store so long metadata string for a matrix with
several elements.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149532602
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/linalg/JsonMatrixConverter.scala ---
@@ -0,0 +1,79 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.linalg
+
+import org.json4s.DefaultFormats
+import org.json4s.JsonDSL._
+import org.json4s.jackson.JsonMethods.{compact, parse => parseJson, render}
+
+private[ml] object JsonMatrixConverter {
+
+ /** Unique class name for identifying JSON object encoded by this class.
*/
+ val className = "org.apache.spark.ml.linalg.Matrix"
--- End diff --
Or can we just use ```type``` to identify vector and matrix? For example,
```type``` less than 10 is reserved for vector and more than 10 is for matrix.
What do you think of it?
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19525: [SPARK-22289] [ML] Add JSON support for Matrix pa...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19525#discussion_r149534129
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/classification/LogisticRegressionSuite.scala
---
@@ -2769,6 +2769,20 @@ class LogisticRegressionSuite
LogisticRegressionSuite.allParamSettings, checkModelData)
}
+ test("read/write with BoundsOnCoefficients") {
+def checkModelData(model: LogisticRegressionModel, model2:
LogisticRegressionModel): Unit = {
+ assert(model.getLowerBoundsOnCoefficients ===
model2.getLowerBoundsOnCoefficients)
+ assert(model.getUpperBoundsOnCoefficients ===
model2.getUpperBoundsOnCoefficients)
--- End diff --
Or we can merge this test case with existing read/write test.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19525: [SPARK-22289] [ML] Add JSON support for Matrix parameter...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19525 Will make a pass soon. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interface of d...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19156 I'd like to make a pass soon. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 @hhbyyh I have compared this implementation with sklearn ```HuberRegressor``` on several dataset listed at https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/regression.html, they can produce consistent result. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149251282
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala
---
@@ -998,6 +1047,198 @@ class LinearRegressionSuite
}
}
}
+
+ test("linear regression (huber loss) with intercept without
regularization") {
+val trainer1 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(true).setStandardization(true)
+val trainer2 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(true).setStandardization(false)
+
+val model1 = trainer1.fit(datasetWithOutlier)
+val model2 = trainer2.fit(datasetWithOutlier)
+
+/*
+ Using the following Python code to load the data and train the model
using
+ scikit-learn package.
+
+ import pandas as pd
+ import numpy as np
+ from sklearn.linear_model import HuberRegressor
+ df = pd.read_csv("path", header = None)
+ X = df[df.columns[1:3]]
+ y = np.array(df[df.columns[0]])
+ huber = HuberRegressor(fit_intercept=True, alpha=0.0, max_iter=100,
epsilon=1.35)
+ huber.fit(X, y)
+
+ >>> huber.coef_
+ array([ 4.68998007, 7.19429011])
+ >>> huber.intercept_
+ 6.3002404351083037
+ >>> huber.scale_
+ 0.077810159205220747
+ */
+val coefficientsPy = Vectors.dense(4.68998007, 7.19429011)
+val interceptPy = 6.30024044
+val scalePy = 0.07781016
+
+assert(model1.coefficients ~= coefficientsPy relTol 1E-3)
+assert(model1.intercept ~== interceptPy relTol 1E-3)
+assert(model1.scale ~== scalePy relTol 1E-3)
+
+// Without regularization, with or without standardization will
converge to the same solution.
+assert(model2.coefficients ~= coefficientsPy relTol 1E-3)
+assert(model2.intercept ~== interceptPy relTol 1E-3)
+assert(model2.scale ~== scalePy relTol 1E-3)
+ }
+
+ test("linear regression (huber loss) without intercept without
regularization") {
+val trainer1 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(false).setStandardization(true)
+val trainer2 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(false).setStandardization(false)
+
+val model1 = trainer1.fit(datasetWithOutlier)
+val model2 = trainer2.fit(datasetWithOutlier)
+
+/*
+ huber = HuberRegressor(fit_intercept=False, alpha=0.0, max_iter=100,
epsilon=1.35)
+ huber.fit(X, y)
+
+ >>> huber.coef_
+ array([ 6.71756703, 5.08873222])
+ >>> huber.intercept_
+ 0.0
+ >>> huber.scale_
+ 2.5560209922722317
+ */
+val coefficientsPy = Vectors.dense(6.71756703, 5.08873222)
+val interceptPy = 0.0
+val scalePy = 2.55602099
+
+assert(model1.coefficients ~= coefficientsPy relTol 1E-3)
+assert(model1.intercept === interceptPy)
+assert(model1.scale ~== scalePy relTol 1E-3)
+
+// Without regularization, with or without standardization will
converge to the same solution.
+assert(model2.coefficients ~= coefficientsPy relTol 1E-3)
+assert(model2.intercept === interceptPy)
+assert(model2.scale ~== scalePy relTol 1E-3)
+ }
+
+ test("linear regression (huber loss) with intercept with L2
regularization") {
+val trainer1 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(true).setRegParam(0.21).setStandardization(true)
+val trainer2 = (new LinearRegression).setLoss("huber")
+ .setFitIntercept(true).setRegParam(0.21).setStandardization(false)
+
+val model1 = trainer1.fit(datasetWithOutlier)
+val model2 = trainer2.fit(datasetWithOutlier)
+
+/*
+ Since scikit-learn HuberRegressor does not support standardization,
+ we do it manually out of the estimator.
+
+ xStd = np.std(X, axis=0)
+ scaledX = X / xStd
+ huber = HuberRegressor(fit_intercept=True, alpha=210, max_iter=100,
epsilon=1.35)
+ huber.fit(scaledX, y)
+
+ >>> np.array(huber.coef_ / xStd)
+ array([ 1.97732633, 3.38816722])
+ >>> huber.intercept_
+ 3.7527581430531227
+ >>> huber.scale_
+ 3.787363673371801
+ */
+val coefficientsPy1 = Vectors.dense(1.97732633, 3.38816722)
+val interceptPy1 = 3.75275814
+val scalePy1 = 3.78736367
+
+assert(model1.coefficients ~= coefficientsPy1 r
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149251168
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -480,10 +638,14 @@ object LinearRegression extends
DefaultParamsReadable[LinearRegression] {
class LinearRegressionModel private[ml] (
@Since("1.4.0") override val uid: String,
@Since("2.0.0") val coefficients: Vector,
-@Since("1.3.0") val intercept: Double)
+@Since("1.3.0") val intercept: Double,
+@Since("2.3.0") val scale: Double)
--- End diff --
This is suggested by @jkbradley , and I also agree that it should be useful
for model interpretation and debugging. And I have provided another constructor
for linear regression model produced by _squared error_.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149250515
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,141 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * Art B. Owen (2006), A robust hybrid of lasso and ridge regression.
+ * (http://statweb.stanford.edu/~owen/reports/hhu.pdf)
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\alpha {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency.
+ *
+ * @param fitIntercept Whether to fit an intercept term.
+ * @param m The shape parameter to control the amount of robustness.
+ * @param bcFeaturesStd The broadcast standard deviation values of the
features.
+ * @param bcParameters including three parts: the regression coefficients
corresponding
+ * to the features, the intercept (if fitIntercept is
ture)
+ * and the scale parameter (sigma).
+ */
+private[ml] class HuberAggregator(
+fitIntercept: Boolean,
+m: Double,
+bcFeaturesStd: Broadcast[Array[Double]])(bcParameters:
Broadcast[Vector])
+ extends DifferentiableLossAggregator[Instance, HuberAggregator] {
+
+ protected override val dim: Int = bcParameters.value.size
+ private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1
+
+ @transient private lazy val coefficients: Array[Double] =
+bcParameters.value.toArray.slice(0, numFeatures)
+ private val sigma: Double = bcParameters.value(dim - 1)
+
+ @transient private lazy val featuresStd = bcFeaturesStd.value
+
+ /**
+ * Add a new training instance to this HuberAggregator, and update the
loss and gradient
+ * of the objective function.
+ *
+ * @param instance The instance of data point to be added.
+ * @return This HuberAggregator object.
+ */
+ def add(instance: Instance): HuberAggregator = {
+instance match { case Instance(label, weight, features) =>
+ require(numFeatures == features.size, s"Dimensions mismatch when
adding new sample." +
+s" Expecting $numFeatures but got ${features.size}.")
+ require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0")
+
+ if (weight == 0.0) return this
+
+ val margin = {
+var sum = 0.0
+features.foreachActive { (index, value) =>
+ if (featuresStd(index) != 0.0 && value != 0.0) {
+sum += coefficients(index) * (value / featuresStd(index))
+ }
+}
+if (fitIntercept) sum += bcParameters.value(dim - 2)
--- End diff --
I created a class variable for ```intercept```.
---
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149250222
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -344,33 +449,58 @@ class LinearRegression @Since("1.3.0")
(@Since("1.3.0") override val uid: String
} else {
None
}
-val costFun = new RDDLossFunction(instances, getAggregatorFunc,
regularization,
- $(aggregationDepth))
-val optimizer = if ($(elasticNetParam) == 0.0 || effectiveRegParam ==
0.0) {
- new BreezeLBFGS[BDV[Double]]($(maxIter), 10, $(tol))
-} else {
- val standardizationParam = $(standardization)
- def effectiveL1RegFun = (index: Int) => {
-if (standardizationParam) {
- effectiveL1RegParam
+val costFun = $(loss) match {
+ case SquaredError =>
+val getAggregatorFunc = new LeastSquaresAggregator(yStd, yMean,
$(fitIntercept),
+ bcFeaturesStd, bcFeaturesMean)(_)
+new RDDLossFunction(instances, getAggregatorFunc, regularization,
$(aggregationDepth))
+ case Huber =>
+val getAggregatorFunc = new HuberAggregator($(fitIntercept),
$(epsilon), bcFeaturesStd)(_)
+new RDDLossFunction(instances, getAggregatorFunc, regularization,
$(aggregationDepth))
+}
+
+val optimizer = $(loss) match {
+ case SquaredError =>
+if ($(elasticNetParam) == 0.0 || effectiveRegParam == 0.0) {
+ new BreezeLBFGS[BDV[Double]]($(maxIter), 10, $(tol))
} else {
- // If `standardization` is false, we still standardize the data
- // to improve the rate of convergence; as a result, we have to
- // perform this reverse standardization by penalizing each
component
- // differently to get effectively the same objective function
when
- // the training dataset is not standardized.
- if (featuresStd(index) != 0.0) effectiveL1RegParam /
featuresStd(index) else 0.0
+ val standardizationParam = $(standardization)
+ def effectiveL1RegFun = (index: Int) => {
+if (standardizationParam) {
+ effectiveL1RegParam
+} else {
+ // If `standardization` is false, we still standardize the
data
+ // to improve the rate of convergence; as a result, we have
to
+ // perform this reverse standardization by penalizing each
component
+ // differently to get effectively the same objective
function when
+ // the training dataset is not standardized.
+ if (featuresStd(index) != 0.0) effectiveL1RegParam /
featuresStd(index) else 0.0
+}
+ }
+ new BreezeOWLQN[Int, BDV[Double]]($(maxIter), 10,
effectiveL1RegFun, $(tol))
}
- }
- new BreezeOWLQN[Int, BDV[Double]]($(maxIter), 10, effectiveL1RegFun,
$(tol))
+ case Huber =>
+val dim = if ($(fitIntercept)) numFeatures + 2 else numFeatures + 1
+val lowerBounds = BDV[Double](Array.fill(dim)(Double.MinValue))
+// Optimize huber loss in space "\sigma > 0"
+lowerBounds(dim - 1) = Double.MinPositiveValue
+val upperBounds = BDV[Double](Array.fill(dim)(Double.MaxValue))
+new BreezeLBFGSB(lowerBounds, upperBounds, $(maxIter), 10, $(tol))
+}
+
+val initialValues = $(loss) match {
+ case SquaredError =>
+Vectors.zeros(numFeatures)
+ case Huber =>
+val dim = if ($(fitIntercept)) numFeatures + 2 else numFeatures + 1
+Vectors.dense(Array.fill(dim)(1.0))
--- End diff --
I don't have reference, just because the initial ```scale``` value should
be great than 0.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247649
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,145 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>Art B. Owen
(2006),
+ * A robust hybrid of lasso and ridge regression.
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\lambda {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency
+ * for normally distributed data. Please refer to chapter 2 of
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>
+ * A robust hybrid of lasso and ridge regression for more detail.
+ *
+ * @param fitIntercept Whether to fit an intercept term.
+ * @param epsilon The shape parameter to control the amount of robustness.
+ * @param bcFeaturesStd The broadcast standard deviation values of the
features.
+ * @param bcParameters including three parts: the regression coefficients
corresponding
+ * to the features, the intercept (if fitIntercept is
ture)
+ * and the scale parameter (sigma).
+ */
+private[ml] class HuberAggregator(
+fitIntercept: Boolean,
+epsilon: Double,
+bcFeaturesStd: Broadcast[Array[Double]])(bcParameters:
Broadcast[Vector])
+ extends DifferentiableLossAggregator[Instance, HuberAggregator] {
+
+ protected override val dim: Int = bcParameters.value.size
+ private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1
+
+ @transient private lazy val coefficients: Array[Double] =
+bcParameters.value.toArray.slice(0, numFeatures)
+ private val sigma: Double = bcParameters.value(dim - 1)
+
+ @transient private lazy val featuresStd = bcFeaturesStd.value
+
+ /**
+ * Add a new training instance to this HuberAggregator, and update the
loss and gradient
+ * of the objective function.
+ *
+ * @param instance The instance of data point to be added.
+ * @return This HuberAggregator object.
+ */
+ def add(instance: Instance): HuberAggregator = {
+instance match { case Instance(label, weight, features) =>
+ require(numFeatures == features.size, s"Dimensions mismatch when
adding new sample." +
+s" Expecting $numFeatures but got ${features.size}.")
+ require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0")
+
+ if (weight == 0.0) return this
+
+ val margin = {
+var sum = 0.0
+features.foreachActive { (index, value) =>
+ if (featuresStd(index) != 0.0 && value != 0.0) {
+sum += coefficients(index) * (value / fe
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247683
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -142,6 +221,9 @@ class LinearRegression @Since("1.3.0") (@Since("1.3.0")
override val uid: String
* For alpha in (0,1), the penalty is a combination of L1 and L2.
* Default is 0.0 which is an L2 penalty.
*
+ * Note: Fitting with huber loss only supports L2 regularization,
--- End diff --
Done.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247570
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,145 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>Art B. Owen
(2006),
+ * A robust hybrid of lasso and ridge regression.
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\lambda {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency
+ * for normally distributed data. Please refer to chapter 2 of
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>
+ * A robust hybrid of lasso and ridge regression for more detail.
+ *
+ * @param fitIntercept Whether to fit an intercept term.
+ * @param epsilon The shape parameter to control the amount of robustness.
+ * @param bcFeaturesStd The broadcast standard deviation values of the
features.
+ * @param bcParameters including three parts: the regression coefficients
corresponding
+ * to the features, the intercept (if fitIntercept is
ture)
+ * and the scale parameter (sigma).
+ */
+private[ml] class HuberAggregator(
+fitIntercept: Boolean,
+epsilon: Double,
+bcFeaturesStd: Broadcast[Array[Double]])(bcParameters:
Broadcast[Vector])
+ extends DifferentiableLossAggregator[Instance, HuberAggregator] {
+
+ protected override val dim: Int = bcParameters.value.size
+ private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1
+
+ @transient private lazy val coefficients: Array[Double] =
+bcParameters.value.toArray.slice(0, numFeatures)
+ private val sigma: Double = bcParameters.value(dim - 1)
+
+ @transient private lazy val featuresStd = bcFeaturesStd.value
+
+ /**
+ * Add a new training instance to this HuberAggregator, and update the
loss and gradient
+ * of the objective function.
+ *
+ * @param instance The instance of data point to be added.
+ * @return This HuberAggregator object.
+ */
+ def add(instance: Instance): HuberAggregator = {
+instance match { case Instance(label, weight, features) =>
+ require(numFeatures == features.size, s"Dimensions mismatch when
adding new sample." +
+s" Expecting $numFeatures but got ${features.size}.")
+ require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0")
+
+ if (weight == 0.0) return this
+
+ val margin = {
+var sum = 0.0
+features.foreachActive { (index, value) =>
+ if (featuresStd(index) != 0.0 && value != 0.0) {
--- End diff --
Done.
---
--
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247543
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,145 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>Art B. Owen
(2006),
+ * A robust hybrid of lasso and ridge regression.
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\lambda {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency
+ * for normally distributed data. Please refer to chapter 2 of
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>
+ * A robust hybrid of lasso and ridge regression for more detail.
--- End diff --
Done.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247240
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -208,6 +292,26 @@ class LinearRegression @Since("1.3.0")
(@Since("1.3.0") override val uid: String
def setAggregationDepth(value: Int): this.type = set(aggregationDepth,
value)
setDefault(aggregationDepth -> 2)
+ /**
+ * Sets the value of param [[loss]].
+ * Default is "squaredError".
+ *
+ * @group setParam
+ */
+ @Since("2.3.0")
+ def setLoss(value: String): this.type = set(loss, value)
+ setDefault(loss -> SquaredError)
+
+ /**
+ * Sets the value of param [[epsilon]].
+ * Default is 1.35.
+ *
+ * @group setExpertParam
+ */
+ @Since("2.3.0")
+ def setEpsilon(value: Double): this.type = set(epsilon, value)
--- End diff --
Document them at the definition of ```epsilon```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247162
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -744,11 +754,20 @@ object LinearRegressionModel extends
MLReadable[LinearRegressionModel] {
val dataPath = new Path(path, "data").toString
val data = sparkSession.read.format("parquet").load(dataPath)
- val Row(intercept: Double, coefficients: Vector) =
-MLUtils.convertVectorColumnsToML(data, "coefficients")
- .select("intercept", "coefficients")
- .head()
- val model = new LinearRegressionModel(metadata.uid, coefficients,
intercept)
+ val (majorVersion, minorVersion) =
majorMinorVersion(metadata.sparkVersion)
+ val model = if (majorVersion < 2 || (majorVersion == 2 &&
minorVersion <= 2)) {
+// Spark 2.2 and before
+val Row(intercept: Double, coefficients: Vector) =
+ MLUtils.convertVectorColumnsToML(data, "coefficients")
+.select("intercept", "coefficients")
+.head()
+new LinearRegressionModel(metadata.uid, coefficients, intercept)
+ } else {
+// Spark 2.3 and later
+val Row(intercept: Double, coefficients: Vector, scale: Double) =
+ data.select("intercept", "coefficients", "scale").head()
+new LinearRegressionModel(metadata.uid, coefficients, intercept,
scale)
+ }
--- End diff --
Of course, I have offline test.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r149247042
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,145 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>Art B. Owen
(2006),
+ * A robust hybrid of lasso and ridge regression.
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\lambda {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency
+ * for normally distributed data. Please refer to chapter 2 of
+ * http://statweb.stanford.edu/~owen/reports/hhu.pdf;>
+ * A robust hybrid of lasso and ridge regression for more detail.
+ *
+ * @param fitIntercept Whether to fit an intercept term.
+ * @param epsilon The shape parameter to control the amount of robustness.
--- End diff --
I have documented them at the definition of ```epsilon``` param in
```LinearRegression```, as there should be public and here is for internal use
only.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19648: [SPARK-14516][ML][FOLLOW-UP] Move ClusteringEvalu...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19648#discussion_r148861446
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/evaluation/ClusteringEvaluatorSuite.scala
---
@@ -22,15 +22,21 @@ import org.apache.spark.ml.param.ParamsSuite
import org.apache.spark.ml.util.DefaultReadWriteTest
import org.apache.spark.ml.util.TestingUtils._
import org.apache.spark.mllib.util.MLlibTestSparkContext
-import org.apache.spark.sql.{DataFrame, SparkSession}
-import org.apache.spark.sql.types.IntegerType
+import org.apache.spark.sql.Dataset
class ClusteringEvaluatorSuite
extends SparkFunSuite with MLlibTestSparkContext with
DefaultReadWriteTest {
import testImplicits._
+ @transient var irisDataset: Dataset[_] = _
--- End diff --
Either is ok, no difference on performance, we just keep consistent with
other test suites.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19648: [SPARK-14516][ML][FOLLOW-UP] Move ClusteringEvaluatorSui...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19648 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with the im...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18538 @jkbradley @mgaido91 I just sent #19648 to move test data to data/mllib, please feel free to review it. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19648: [SPARK-14516][ML][FOLLOW-UP] Move ClusteringEvalu...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/19648 [SPARK-14516][ML][FOLLOW-UP] Move ClusteringEvaluatorSuite test data to data/mllib. ## What changes were proposed in this pull request? Move ```ClusteringEvaluatorSuite``` test data(iris) to data/mllib, to prevent from re-creating a new folder. ## How was this patch tested? Existing tests. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark spark-14516 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19648.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19648 commit 0fd9a5c617053ced4210432f261a0053a04442dd Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-11-03T01:03:23Z Move ClusteringEvaluatorSuite test data to data/mllib. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with the im...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18538 @mgaido91 Don't worry, I'll post a follow-up PR for discussion in a few days. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
GitHub user yanboliang reopened a pull request: https://github.com/apache/spark/pull/19020 [SPARK-3181] [ML] Implement huber loss for LinearRegression. ## What changes were proposed in this pull request? MLlib ```LinearRegression``` supports _huber_ loss addition to _leastSquares_ loss. The huber loss objective function is: 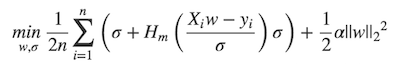 Refer Eq.(6) and Eq.(8) in [A robust hybrid of lasso and ridge regression](http://statweb.stanford.edu/~owen/reports/hhu.pdf). This objective is jointly convex as a function of (w, Ï) â R Ã (0,â), we can use L-BFGS-B to solve it. The current implementation is a straight forward porting for Python scikit-learn [```HuberRegressor```](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.HuberRegressor.html). There are some differences: * We use mean loss (```lossSum/weightSum```), but sklearn uses total loss (```lossSum```). * We multiply the loss function and L2 regularization by 1/2. It does not affect the result if we multiply the whole formula by a factor, we just keep consistent with _leastSquares_ loss. So if fitting w/o regularization, MLlib and sklearn produce the same output. If fitting w/ regularization, MLlib should set ```regParam``` divide by the number of instances to match the output of sklearn. ## How was this patch tested? Unit tests. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark spark-3181 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19020.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19020 commit c208c7b3ac4917098dd07ddc777e65fae77e21d0 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-20T05:45:36Z Implement HuberAggregator and add tests. commit d72059debf079bb5aebfb9010e1a25241dc82856 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-21T13:43:28Z Implement huber loss for LinearRegression. commit c7456837166c70aedb2c164b7e5f17ebd7033c56 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T03:21:42Z Update HuberAggregator and tests. commit 43822bde3983214c54a571c86fbc534170b86415 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T04:34:02Z Update params doc and check for illegal params. commit 5b431461acd15284f635da5f7c220f186386e351 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T07:25:27Z Update LinearRegression test suites. commit 9545cdef56251755669c6fd4dbf301779d4115f3 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T08:19:29Z Add mima excludes. commit f8fb60ace2cfb1e2cf739f09dab01e60ba7cb4df Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T10:29:16Z Fix docs. commit 0951b02e15632200bb0e3052f95ec5126091f98e Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-08-22T11:25:39Z Fix annotation. commit 8bdd625c996dca93ca915446cc4b3ff39b7478e9 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-01T07:06:00Z Update and reorg test cases. commit ae2a9f89d7d88599e06709b15e3f368692b1e067 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-01T07:26:28Z Minor update for tests. commit d21b4fb1397c48e43dcb9dc13344788d8caa1036 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-01T08:07:46Z Rename m to epsilon. commit 3d3f1ec6696a1cd6f3cd2ee1bfb4e74790403c84 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-22T07:41:25Z Address review comments. commit 7359635e9a2a4f050418b5d3f51ee85fb73b4d2d Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-22T08:34:20Z Add loss function formula for LinearRegression. commit 8c6622f68ea81cedbeb3f03f957b335a99dedd46 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-10-03T05:16:26Z Expose scale for LinearRegressionModel. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang closed the pull request at: https://github.com/apache/spark/pull/19020 --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #16548: [SPARK-19158][SPARKR][EXAMPLES] Fix ml.R example fails d...
Github user yanboliang commented on the issue:
https://github.com/apache/spark/pull/16548
@holdenk Could you let me know where we meet similar issue in the
fulltests? AFAIK, we test functions in ```e1071``` only when it was installed
on that node, like following:
```
if (requireNamespace("e1071", quietly = TRUE)) {
expect_error(m <- e1071::naiveBayes(Survived ~ ., data = t1), NA)
expect_equal(as.character(predict(m, t1[1, ])), "Yes")
}
```
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 @sethah To the issue that whether huber linear regression share codebase with ```LinearRegression```, we have discussion at [JIRA](https://issues.apache.org/jira/browse/SPARK-3181). At last @dbtsai and I reached an agreement to combine them in a single class. Actually in sklearn, there are separate [```HuberRegressor```](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.HuberRegressor.html) and [```SGDRegressor```](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.SGDRegressor.html) support to train linear regression with huber loss. Also, in this [paper](http://statweb.stanford.edu/~owen/reports/hhu.pdf), huber regression is a robust hybrid of lasso and ridge regression, so I think we can combine them together. @jkbradley @MLnick @WeichenXu123 @hhbyyh What's your opinion? Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19020: [SPARK-3181] [ML] Implement huber loss for LinearRegress...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19020 @jkbradley Thanks for your comments, I have addressed all your inline comments. Please see replies to your other questions below: > Echoing @WeichenXu123 's comment: Why use "epsilon" as the Param name? We have two candidate name: _epsilon_ or _m_ , both of them are not very descriptive. I referred sklearn [HuberRegressor](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.HuberRegressor.html), and keep consistent with it. > I'd like us to provide the estimated scaling factor (sigma from the paper) in the Model. That seems useful for model interpretation and debugging. I'm hesitating to add it to ```LinearRegression``` in case to confuse users who just try with different losses, but I'm OK to add it. Which should be output for _sigma_ if users fit with _squaredError_? A default value or throwing exception? Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140440076
--- Diff:
mllib/src/test/scala/org/apache/spark/ml/regression/LinearRegressionSuite.scala
---
@@ -998,6 +1047,172 @@ class LinearRegressionSuite
}
}
}
+
+ test("linear regression (huber loss) with intercept without
regularization") {
--- End diff --
Yes, the test data was composed by two parts: inlierData and outlierData,
and I have checked both regimes have been test. Thanks.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140439435
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -220,12 +283,12 @@ class LinearRegression @Since("1.3.0")
(@Since("1.3.0") override val uid: String
}
val instr = Instrumentation.create(this, dataset)
-instr.logParams(labelCol, featuresCol, weightCol, predictionCol,
solver, tol,
- elasticNetParam, fitIntercept, maxIter, regParam, standardization,
aggregationDepth)
+instr.logParams(labelCol, featuresCol, weightCol, predictionCol,
solver, tol, elasticNetParam,
--- End diff --
Done.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140439369
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -69,19 +69,57 @@ private[regression] trait LinearRegressionParams
extends PredictorParams
"The solver algorithm for optimization. Supported options: " +
s"${supportedSolvers.mkString(", ")}. (Default auto)",
ParamValidators.inArray[String](supportedSolvers))
+
+ /**
+ * The loss function to be optimized.
+ * Supported options: "leastSquares" and "huber".
+ * Default: "leastSquares"
+ *
+ * @group param
+ */
+ @Since("2.3.0")
+ final override val loss: Param[String] = new Param[String](this, "loss",
"The loss function to" +
+s" be optimized. Supported options: ${supportedLosses.mkString(", ")}.
(Default leastSquares)",
+ParamValidators.inArray[String](supportedLosses))
+
+ /**
+ * The shape parameter to control the amount of robustness. Must be
1.0.
+ * At larger values of epsilon, the huber criterion becomes more similar
to least squares
+ * regression; for small values of epsilon, the criterion is more
similar to L1 regression.
+ * Default is 1.35 to get as much robustness as possible while retaining
+ * 95% statistical efficiency for normally distributed data.
+ * Only valid when "loss" is "huber".
+ */
+ @Since("2.3.0")
+ final val epsilon = new DoubleParam(this, "epsilon", "The shape
parameter to control the " +
+"amount of robustness. Must be > 1.0.", ParamValidators.gt(1.0))
+
+ /** @group getParam */
+ @Since("2.3.0")
+ def getEpsilon: Double = $(epsilon)
+
+ override protected def validateAndTransformSchema(
+ schema: StructType,
+ fitting: Boolean,
+ featuresDataType: DataType): StructType = {
+if ($(loss) == Huber) {
+ require($(solver)!= Normal, "LinearRegression with huber loss
doesn't support " +
+"normal solver, please change solver to auto or l-bfgs.")
+ require($(elasticNetParam) == 0.0, "LinearRegression with huber loss
only supports " +
+s"L2 regularization, but got elasticNetParam =
$getElasticNetParam.")
+
+}
+super.validateAndTransformSchema(schema, fitting, featuresDataType)
+ }
}
/**
* Linear regression.
*
- * The learning objective is to minimize the squared error, with
regularization.
- * The specific squared error loss function used is:
- *
- *
- *$$
- *L = 1/2n ||A coefficients - y||^2^
- *$$
- *
+ * The learning objective is to minimize the specified loss function, with
regularization.
+ * This supports two loss functions:
+ * - leastSquares (a.k.a squared loss)
--- End diff --
I agree, and I added math formula for both _squaredError_ and _huber_ loss
function.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140439119
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,142 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * Art B. Owen (2006), A robust hybrid of lasso and ridge regression.
+ * (http://statweb.stanford.edu/~owen/reports/hhu.pdf)
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\alpha {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency.
--- End diff --
Done.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140439171
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/regression/LinearRegression.scala ---
@@ -69,19 +69,57 @@ private[regression] trait LinearRegressionParams
extends PredictorParams
"The solver algorithm for optimization. Supported options: " +
s"${supportedSolvers.mkString(", ")}. (Default auto)",
ParamValidators.inArray[String](supportedSolvers))
+
+ /**
+ * The loss function to be optimized.
+ * Supported options: "leastSquares" and "huber".
+ * Default: "leastSquares"
+ *
+ * @group param
+ */
+ @Since("2.3.0")
+ final override val loss: Param[String] = new Param[String](this, "loss",
"The loss function to" +
+s" be optimized. Supported options: ${supportedLosses.mkString(", ")}.
(Default leastSquares)",
+ParamValidators.inArray[String](supportedLosses))
+
+ /**
+ * The shape parameter to control the amount of robustness. Must be
1.0.
+ * At larger values of epsilon, the huber criterion becomes more similar
to least squares
+ * regression; for small values of epsilon, the criterion is more
similar to L1 regression.
+ * Default is 1.35 to get as much robustness as possible while retaining
+ * 95% statistical efficiency for normally distributed data.
+ * Only valid when "loss" is "huber".
+ */
+ @Since("2.3.0")
+ final val epsilon = new DoubleParam(this, "epsilon", "The shape
parameter to control the " +
--- End diff --
Done.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19020: [SPARK-3181] [ML] Implement huber loss for Linear...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19020#discussion_r140439140
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
---
@@ -0,0 +1,142 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+package org.apache.spark.ml.optim.aggregator
+
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.feature.Instance
+import org.apache.spark.ml.linalg.Vector
+
+/**
+ * HuberAggregator computes the gradient and loss for a huber loss
function,
+ * as used in robust regression for samples in sparse or dense vector in
an online fashion.
+ *
+ * The huber loss function based on:
+ * Art B. Owen (2006), A robust hybrid of lasso and ridge regression.
+ * (http://statweb.stanford.edu/~owen/reports/hhu.pdf)
+ *
+ * Two HuberAggregator can be merged together to have a summary of loss
and gradient of
+ * the corresponding joint dataset.
+ *
+ * The huber loss function is given by
+ *
+ *
+ * $$
+ * \begin{align}
+ * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma +
+ * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) +
\frac{1}{2}\alpha {||w||_2}^2}
+ * \end{align}
+ * $$
+ *
+ *
+ * where
+ *
+ *
+ * $$
+ * \begin{align}
+ * H_m(z) = \begin{cases}
+ *z^2, & \text {if } |z| \epsilon, \\
+ *2\epsilon|z| - \epsilon^2, & \text{otherwise}
+ *\end{cases}
+ * \end{align}
+ * $$
+ *
+ *
+ * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95%
statistical efficiency.
+ *
+ * @param fitIntercept Whether to fit an intercept term.
+ * @param epsilon The shape parameter to control the amount of robustness.
+ * @param bcFeaturesStd The broadcast standard deviation values of the
features.
+ * @param bcParameters including three parts: the regression coefficients
corresponding
+ * to the features, the intercept (if fitIntercept is
ture)
+ * and the scale parameter (sigma).
+ */
+private[ml] class HuberAggregator(
+fitIntercept: Boolean,
+epsilon: Double,
+bcFeaturesStd: Broadcast[Array[Double]])(bcParameters:
Broadcast[Vector])
+ extends DifferentiableLossAggregator[Instance, HuberAggregator] {
+
+ protected override val dim: Int = bcParameters.value.size
+ private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1
+
+ @transient private lazy val coefficients: Array[Double] =
+bcParameters.value.toArray.slice(0, numFeatures)
+ private val sigma: Double = bcParameters.value(dim - 1)
+
+ @transient private lazy val featuresStd = bcFeaturesStd.value
+
+ /**
+ * Add a new training instance to this HuberAggregator, and update the
loss and gradient
+ * of the objective function.
+ *
+ * @param instance The instance of data point to be added.
+ * @return This HuberAggregator object.
+ */
+ def add(instance: Instance): HuberAggregator = {
+instance match { case Instance(label, weight, features) =>
+ require(numFeatures == features.size, s"Dimensions mismatch when
adding new sample." +
+s" Expecting $numFeatures but got ${features.size}.")
+ require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0")
+
+ if (weight == 0.0) return this
+
+ val margin = {
+var sum = 0.0
+features.foreachActive { (index, value) =>
+ if (featuresStd(index) != 0.0 && value != 0.0) {
+sum += coefficients(index) * (value / featuresStd(index))
+ }
+}
+if (fitIntercept) sum += bcParameters.value(dim - 2)
+sum
+ }
+ val linearLoss = label - margin
+
+ if (math.abs(l
[GitHub] spark issue #19204: [SPARK-21981][PYTHON][ML] Added Python interface for Clu...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19204 Merged into master, thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19204#discussion_r139718610 --- Diff: python/pyspark/ml/evaluation.py --- @@ -328,6 +329,77 @@ def setParams(self, predictionCol="prediction", labelCol="label", kwargs = self._input_kwargs return self._set(**kwargs) + +@inherit_doc +class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol, + JavaMLReadable, JavaMLWritable): +""" +.. note:: Experimental + +Evaluator for Clustering results, which expects two input +columns: prediction and features. + +>>> from pyspark.ml.linalg import Vectors +>>> scoreAndLabels = map(lambda x: (Vectors.dense(x[0]), x[1]), --- End diff -- ```scoreAndLabels``` -> ```featureAndPredictions```, the dataset here is different from other evaluators, we should use more accurate name. Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19156: [SPARK-19634][SQL][ML][FOLLOW-UP] Improve interface of d...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19156 @WeichenXu123 Sorry for late response, really busy in these days. I will take a look in a few days. Thanks for your patience. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19262: [MINOR][ML] Remove unnecessary default value setting for...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19262 Merged into master. Thanks for reviewing. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19204#discussion_r139312695
--- Diff: python/pyspark/ml/evaluation.py ---
@@ -328,6 +329,86 @@ def setParams(self, predictionCol="prediction",
labelCol="label",
kwargs = self._input_kwargs
return self._set(**kwargs)
+
+@inherit_doc
+class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol,
+ JavaMLReadable, JavaMLWritable):
+"""
+.. note:: Experimental
+
+Evaluator for Clustering results, which expects two input
+columns: prediction and features.
+
+>>> from sklearn import datasets
+>>> from pyspark.sql.types import *
+>>> from pyspark.ml.linalg import Vectors, VectorUDT
+>>> from pyspark.ml.evaluation import ClusteringEvaluator
+...
+>>> iris = datasets.load_iris()
+>>> iris_rows = [(Vectors.dense(x), int(iris.target[i]))
+... for i, x in enumerate(iris.data)]
+>>> schema = StructType([
+...StructField("features", VectorUDT(), True),
+...StructField("cluster_id", IntegerType(), True)])
+>>> rdd = spark.sparkContext.parallelize(iris_rows)
+>>> dataset = spark.createDataFrame(rdd, schema)
+...
+>>> evaluator = ClusteringEvaluator(predictionCol="cluster_id")
+>>> evaluator.evaluate(dataset)
+0.656...
+>>> ce_path = temp_path + "/ce"
+>>> evaluator.save(ce_path)
+>>> evaluator2 = ClusteringEvaluator.load(ce_path)
+>>> str(evaluator2.getPredictionCol())
+'cluster_id'
+
+.. versionadded:: 2.3.0
+"""
+metricName = Param(Params._dummy(), "metricName",
+ "metric name in evaluation (silhouette)",
+ typeConverter=TypeConverters.toString)
+
+@keyword_only
+def __init__(self, predictionCol="prediction", featuresCol="features",
+ metricName="silhouette"):
+"""
+__init__(self, predictionCol="prediction", featuresCol="features",
\
+ metricName="silhouette")
+"""
+super(ClusteringEvaluator, self).__init__()
+self._java_obj = self._new_java_obj(
+"org.apache.spark.ml.evaluation.ClusteringEvaluator", self.uid)
+self._setDefault(predictionCol="prediction",
featuresCol="features",
--- End diff --
I sent #19262 to fix same issue for other evaluators, please feel free to
comment. Thanks.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19262: [MINOR][ML] Remove unnecessary default value sett...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/19262 [MINOR][ML] Remove unnecessary default value setting for evaluators. ## What changes were proposed in this pull request? Remove unnecessary default value setting for all evaluators, as we have set them in corresponding _HasXXX_ base classes. ## How was this patch tested? Existing tests. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark evaluation Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19262.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19262 commit 84c6c9e394939fa97a6d3888547ac269b234a2c0 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-17T14:48:35Z Remove unnecessary default value setting for evaluators. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19204#discussion_r139312388
--- Diff: python/pyspark/ml/evaluation.py ---
@@ -328,6 +329,86 @@ def setParams(self, predictionCol="prediction",
labelCol="label",
kwargs = self._input_kwargs
return self._set(**kwargs)
+
+@inherit_doc
+class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol,
+ JavaMLReadable, JavaMLWritable):
+"""
+.. note:: Experimental
+
+Evaluator for Clustering results, which expects two input
+columns: prediction and features.
+
+>>> from sklearn import datasets
+>>> from pyspark.sql.types import *
+>>> from pyspark.ml.linalg import Vectors, VectorUDT
+>>> from pyspark.ml.evaluation import ClusteringEvaluator
+...
+>>> iris = datasets.load_iris()
+>>> iris_rows = [(Vectors.dense(x), int(iris.target[i]))
+... for i, x in enumerate(iris.data)]
+>>> schema = StructType([
+...StructField("features", VectorUDT(), True),
+...StructField("cluster_id", IntegerType(), True)])
--- End diff --
```cluster_id``` -> ```prediction``` to emphasize this is the prediction
value, not ground truth.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19204#discussion_r139312199
--- Diff: python/pyspark/ml/evaluation.py ---
@@ -328,6 +329,86 @@ def setParams(self, predictionCol="prediction",
labelCol="label",
kwargs = self._input_kwargs
return self._set(**kwargs)
+
+@inherit_doc
+class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol,
+ JavaMLReadable, JavaMLWritable):
+"""
+.. note:: Experimental
+
+Evaluator for Clustering results, which expects two input
+columns: prediction and features.
+
+>>> from sklearn import datasets
+>>> from pyspark.sql.types import *
+>>> from pyspark.ml.linalg import Vectors, VectorUDT
+>>> from pyspark.ml.evaluation import ClusteringEvaluator
+...
+>>> iris = datasets.load_iris()
+>>> iris_rows = [(Vectors.dense(x), int(iris.target[i]))
+... for i, x in enumerate(iris.data)]
+>>> schema = StructType([
+...StructField("features", VectorUDT(), True),
+...StructField("cluster_id", IntegerType(), True)])
+>>> rdd = spark.sparkContext.parallelize(iris_rows)
+>>> dataset = spark.createDataFrame(rdd, schema)
+...
+>>> evaluator = ClusteringEvaluator(predictionCol="cluster_id")
+>>> evaluator.evaluate(dataset)
+0.656...
+>>> ce_path = temp_path + "/ce"
+>>> evaluator.save(ce_path)
+>>> evaluator2 = ClusteringEvaluator.load(ce_path)
+>>> str(evaluator2.getPredictionCol())
+'cluster_id'
+
+.. versionadded:: 2.3.0
+"""
+metricName = Param(Params._dummy(), "metricName",
+ "metric name in evaluation (silhouette)",
+ typeConverter=TypeConverters.toString)
+
+@keyword_only
+def __init__(self, predictionCol="prediction", featuresCol="features",
+ metricName="silhouette"):
+"""
+__init__(self, predictionCol="prediction", featuresCol="features",
\
+ metricName="silhouette")
+"""
+super(ClusteringEvaluator, self).__init__()
+self._java_obj = self._new_java_obj(
+"org.apache.spark.ml.evaluation.ClusteringEvaluator", self.uid)
+self._setDefault(predictionCol="prediction",
featuresCol="features",
--- End diff --
Remove setting default value for ```predictionCol``` and ```featuresCol```,
as they have been set in ```HasPredictionCol``` and ```HasFeaturesCol```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19204#discussion_r139312034 --- Diff: python/pyspark/ml/evaluation.py --- @@ -328,6 +329,86 @@ def setParams(self, predictionCol="prediction", labelCol="label", kwargs = self._input_kwargs return self._set(**kwargs) + +@inherit_doc +class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol, + JavaMLReadable, JavaMLWritable): +""" +.. note:: Experimental + +Evaluator for Clustering results, which expects two input +columns: prediction and features. + +>>> from sklearn import datasets +>>> from pyspark.sql.types import * +>>> from pyspark.ml.linalg import Vectors, VectorUDT +>>> from pyspark.ml.evaluation import ClusteringEvaluator +... +>>> iris = datasets.load_iris() --- End diff -- Please don't involves other libraries if not necessary, here the doc test is used to show how to use ```ClusteringEvaluator``` to fresh users, so we should focus on evaluator and keep it as simple as possible. You can refer other evaluator to construct simple dataset. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19204: [SPARK-21981][PYTHON][ML] Added Python interface ...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19204#discussion_r139312046 --- Diff: python/pyspark/ml/evaluation.py --- @@ -328,6 +329,86 @@ def setParams(self, predictionCol="prediction", labelCol="label", kwargs = self._input_kwargs return self._set(**kwargs) + +@inherit_doc +class ClusteringEvaluator(JavaEvaluator, HasPredictionCol, HasFeaturesCol, + JavaMLReadable, JavaMLWritable): +""" +.. note:: Experimental + +Evaluator for Clustering results, which expects two input +columns: prediction and features. + +>>> from sklearn import datasets +>>> from pyspark.sql.types import * +>>> from pyspark.ml.linalg import Vectors, VectorUDT +>>> from pyspark.ml.evaluation import ClusteringEvaluator --- End diff -- Remove this, it's not necessary. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19220: [SPARK-18608][ML][FOLLOWUP] Fix double caching for PySpa...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19220 Merged into master, thanks for all reviewing. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19185: [Spark-21854] Added LogisticRegressionTrainingSum...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19185#discussion_r138792851 --- Diff: python/pyspark/ml/tests.py --- @@ -1464,20 +1464,79 @@ def test_logistic_regression_summary(self): self.assertEqual(s.probabilityCol, "probability") self.assertEqual(s.labelCol, "label") self.assertEqual(s.featuresCol, "features") +self.assertEqual(s.predictionCol, "prediction") objHist = s.objectiveHistory self.assertTrue(isinstance(objHist, list) and isinstance(objHist[0], float)) self.assertGreater(s.totalIterations, 0) +self.assertTrue(isinstance(s.labels, list)) +self.assertTrue(isinstance(s.truePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.falsePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.precisionByLabel, list)) +self.assertTrue(isinstance(s.recallByLabel, list)) +self.assertTrue(isinstance(s.fMeasureByLabel(), list)) +self.assertTrue(isinstance(s.fMeasureByLabel(1.0), list)) self.assertTrue(isinstance(s.roc, DataFrame)) self.assertAlmostEqual(s.areaUnderROC, 1.0, 2) self.assertTrue(isinstance(s.pr, DataFrame)) self.assertTrue(isinstance(s.fMeasureByThreshold, DataFrame)) self.assertTrue(isinstance(s.precisionByThreshold, DataFrame)) self.assertTrue(isinstance(s.recallByThreshold, DataFrame)) +self.assertAlmostEqual(s.accuracy, 1.0, 2) +self.assertAlmostEqual(s.weightedTruePositiveRate, 1.0, 2) +self.assertAlmostEqual(s.weightedFalsePositiveRate, 0.0, 2) +self.assertAlmostEqual(s.weightedRecall, 1.0, 2) +self.assertAlmostEqual(s.weightedPrecision, 1.0, 2) +self.assertAlmostEqual(s.weightedFMeasure(), 1.0, 2) +self.assertAlmostEqual(s.weightedFMeasure(1.0), 1.0, 2) # test evaluation (with training dataset) produces a summary with same values # one check is enough to verify a summary is returned, Scala version runs full test sameSummary = model.evaluate(df) self.assertAlmostEqual(sameSummary.areaUnderROC, s.areaUnderROC) +def test_multiclass_logistic_regression_summary(self): +df = self.spark.createDataFrame([(1.0, 2.0, Vectors.dense(1.0)), + (0.0, 2.0, Vectors.sparse(1, [], [])), + (2.0, 2.0, Vectors.dense(2.0)), + (2.0, 2.0, Vectors.dense(1.9))], +["label", "weight", "features"]) +lr = LogisticRegression(maxIter=5, regParam=0.01, weightCol="weight", fitIntercept=False) +model = lr.fit(df) +self.assertTrue(model.hasSummary) +s = model.summary +# test that api is callable and returns expected types +self.assertTrue(isinstance(s.predictions, DataFrame)) +self.assertEqual(s.probabilityCol, "probability") +self.assertEqual(s.labelCol, "label") +self.assertEqual(s.featuresCol, "features") +self.assertEqual(s.predictionCol, "prediction") +objHist = s.objectiveHistory +self.assertTrue(isinstance(objHist, list) and isinstance(objHist[0], float)) +self.assertGreater(s.totalIterations, 0) +self.assertTrue(isinstance(s.labels, list)) +self.assertTrue(isinstance(s.truePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.falsePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.precisionByLabel, list)) +self.assertTrue(isinstance(s.recallByLabel, list)) +self.assertTrue(isinstance(s.fMeasureByLabel(), list)) +self.assertTrue(isinstance(s.fMeasureByLabel(1.0), list)) +self.assertAlmostEqual(s.accuracy, 0.75, 2) +self.assertAlmostEqual(s.weightedTruePositiveRate, 0.75, 2) +self.assertAlmostEqual(s.weightedFalsePositiveRate, 0.25, 2) +self.assertAlmostEqual(s.weightedRecall, 0.75, 2) +self.assertAlmostEqual(s.weightedPrecision, 0.583, 2) +self.assertAlmostEqual(s.weightedFMeasure(), 0.65, 2) +self.assertAlmostEqual(s.weightedFMeasure(1.0), 0.65, 2) +# test evaluation (with training dataset) produces a summary with same values +# one check is enough to verify a summary is returned, Scala version runs full test +sameSummary = model.evaluate(df) +self.assertAlmostEqual(sameSummary.accuracy, s.accuracy) --- End diff -- Nit: Like mention
[GitHub] spark issue #19220: [SPARK-18608][ML][FOLLOWUP] Fix double caching for PySpa...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19220 cc @zhengruifeng @jkbradley @WeichenXu123 --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18902: [SPARK-21690][ML] one-pass imputer
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18902 Merged into master. Thanks for all. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19220: [SPARK-18608][ML][FOLLOWUP] Fix double caching fo...
GitHub user yanboliang opened a pull request: https://github.com/apache/spark/pull/19220 [SPARK-18608][ML][FOLLOWUP] Fix double caching for PySpark OneVsRest. ## What changes were proposed in this pull request? #19197 fixed double caching for MLlib algorithms, but missed PySpark ```OneVsRest```, this PR fixed it. ## How was this patch tested? Existing tests. You can merge this pull request into a Git repository by running: $ git pull https://github.com/yanboliang/spark SPARK-18608 Alternatively you can review and apply these changes as the patch at: https://github.com/apache/spark/pull/19220.patch To close this pull request, make a commit to your master/trunk branch with (at least) the following in the commit message: This closes #19220 commit e22ff7fd6d2e203878a61e9835bd854a9f1bbbe4 Author: Yanbo Liang <yblia...@gmail.com> Date: 2017-09-13T11:27:56Z Fix double caching for PySpark OneVsRest. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with the im...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18538 @mgaido91 I opened [SPARK-21981](https://issues.apache.org/jira/browse/SPARK-21981) for Python API, would you like to work on it? Thanks. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with the im...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18538 I'm merging this into master, thanks for all. If anyone has more comments, we can address them in follow-up PRs. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #19185: [Spark-21854] Added LogisticRegressionTrainingSummary fo...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/19185 Jenkins, test this please. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark issue #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with the im...
Github user yanboliang commented on the issue: https://github.com/apache/spark/pull/18538 @mgaido91 These are my last comments, it should be ready to merge once they are addressed. Thanks for your contribution. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/18538#discussion_r138255937
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/evaluation/ClusteringEvaluator.scala
---
@@ -0,0 +1,438 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+
+package org.apache.spark.ml.evaluation
+
+import org.apache.spark.SparkContext
+import org.apache.spark.annotation.{Experimental, Since}
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.linalg.{BLAS, DenseVector, Vector, Vectors,
VectorUDT}
+import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
+import org.apache.spark.ml.param.shared.{HasFeaturesCol, HasPredictionCol}
+import org.apache.spark.ml.util.{DefaultParamsReadable,
DefaultParamsWritable, Identifiable, SchemaUtils}
+import org.apache.spark.sql.{DataFrame, Dataset}
+import org.apache.spark.sql.functions.{avg, col, udf}
+import org.apache.spark.sql.types.IntegerType
+
+/**
+ * :: Experimental ::
+ * Evaluator for clustering results.
+ * The metric computes the Silhouette measure
+ * using the squared Euclidean distance.
+ *
+ * The Silhouette is a measure for the validation
+ * of the consistency within clusters. It ranges
+ * between 1 and -1, where a value close to 1
+ * means that the points in a cluster are close
+ * to the other points in the same cluster and
+ * far from the points of the other clusters.
+ */
+@Experimental
+@Since("2.3.0")
+class ClusteringEvaluator @Since("2.3.0") (@Since("2.3.0") override val
uid: String)
+ extends Evaluator with HasPredictionCol with HasFeaturesCol with
DefaultParamsWritable {
+
+ @Since("2.3.0")
+ def this() = this(Identifiable.randomUID("cluEval"))
+
+ @Since("2.3.0")
+ override def copy(pMap: ParamMap): ClusteringEvaluator =
this.defaultCopy(pMap)
+
+ @Since("2.3.0")
+ override def isLargerBetter: Boolean = true
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setPredictionCol(value: String): this.type = set(predictionCol,
value)
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setFeaturesCol(value: String): this.type = set(featuresCol, value)
+
+ /**
+ * param for metric name in evaluation
+ * (supports `"silhouette"` (default))
+ * @group param
+ */
+ @Since("2.3.0")
+ val metricName: Param[String] = {
+val allowedParams = ParamValidators.inArray(Array("silhouette"))
+new Param(
+ this,
+ "metricName",
+ "metric name in evaluation (silhouette)",
+ allowedParams
+)
+ }
+
+ /** @group getParam */
+ @Since("2.3.0")
+ def getMetricName: String = $(metricName)
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setMetricName(value: String): this.type = set(metricName, value)
+
+ setDefault(metricName -> "silhouette")
+
+ @Since("2.3.0")
+ override def evaluate(dataset: Dataset[_]): Double = {
+SchemaUtils.checkColumnType(dataset.schema, $(featuresCol), new
VectorUDT)
+SchemaUtils.checkColumnType(dataset.schema, $(predictionCol),
IntegerType)
--- End diff --
We should support all numeric type for prediction column, not only integer.
```
SchemaUtils.checkNumericType(schema, $(labelCol))
```
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/18538#discussion_r138256035
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/evaluation/ClusteringEvaluator.scala
---
@@ -0,0 +1,438 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+
+package org.apache.spark.ml.evaluation
+
+import org.apache.spark.SparkContext
+import org.apache.spark.annotation.{Experimental, Since}
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.linalg.{BLAS, DenseVector, Vector, Vectors,
VectorUDT}
+import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
+import org.apache.spark.ml.param.shared.{HasFeaturesCol, HasPredictionCol}
+import org.apache.spark.ml.util.{DefaultParamsReadable,
DefaultParamsWritable, Identifiable, SchemaUtils}
+import org.apache.spark.sql.{DataFrame, Dataset}
+import org.apache.spark.sql.functions.{avg, col, udf}
+import org.apache.spark.sql.types.IntegerType
+
+/**
+ * :: Experimental ::
+ * Evaluator for clustering results.
+ * The metric computes the Silhouette measure
+ * using the squared Euclidean distance.
+ *
+ * The Silhouette is a measure for the validation
+ * of the consistency within clusters. It ranges
+ * between 1 and -1, where a value close to 1
+ * means that the points in a cluster are close
+ * to the other points in the same cluster and
+ * far from the points of the other clusters.
+ */
+@Experimental
+@Since("2.3.0")
+class ClusteringEvaluator @Since("2.3.0") (@Since("2.3.0") override val
uid: String)
+ extends Evaluator with HasPredictionCol with HasFeaturesCol with
DefaultParamsWritable {
+
+ @Since("2.3.0")
+ def this() = this(Identifiable.randomUID("cluEval"))
+
+ @Since("2.3.0")
+ override def copy(pMap: ParamMap): ClusteringEvaluator =
this.defaultCopy(pMap)
+
+ @Since("2.3.0")
+ override def isLargerBetter: Boolean = true
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setPredictionCol(value: String): this.type = set(predictionCol,
value)
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setFeaturesCol(value: String): this.type = set(featuresCol, value)
+
+ /**
+ * param for metric name in evaluation
+ * (supports `"silhouette"` (default))
+ * @group param
+ */
+ @Since("2.3.0")
+ val metricName: Param[String] = {
+val allowedParams = ParamValidators.inArray(Array("silhouette"))
+new Param(
+ this,
+ "metricName",
+ "metric name in evaluation (silhouette)",
+ allowedParams
+)
--- End diff --
Reorg as:
```
val metricName: Param[String] = {
val allowedParams = ParamValidators.inArray(Array("squaredSilhouette"))
new Param(
this, "metricName", "metric name in evaluation (squaredSilhouette)",
allowedParams)
}
```
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/18538#discussion_r138255648
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/evaluation/ClusteringEvaluator.scala
---
@@ -0,0 +1,438 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+
+package org.apache.spark.ml.evaluation
+
+import org.apache.spark.SparkContext
+import org.apache.spark.annotation.{Experimental, Since}
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.linalg.{BLAS, DenseVector, Vector, Vectors,
VectorUDT}
+import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
+import org.apache.spark.ml.param.shared.{HasFeaturesCol, HasPredictionCol}
+import org.apache.spark.ml.util.{DefaultParamsReadable,
DefaultParamsWritable, Identifiable, SchemaUtils}
+import org.apache.spark.sql.{DataFrame, Dataset}
+import org.apache.spark.sql.functions.{avg, col, udf}
+import org.apache.spark.sql.types.IntegerType
+
+/**
+ * :: Experimental ::
--- End diff --
Usually we leave a blank line under ```:: Experimental ::```.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #18538: [SPARK-14516][ML] Adding ClusteringEvaluator with...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/18538#discussion_r138255474
--- Diff:
mllib/src/main/scala/org/apache/spark/ml/evaluation/ClusteringEvaluator.scala
---
@@ -0,0 +1,438 @@
+/*
+ * Licensed to the Apache Software Foundation (ASF) under one or more
+ * contributor license agreements. See the NOTICE file distributed with
+ * this work for additional information regarding copyright ownership.
+ * The ASF licenses this file to You under the Apache License, Version 2.0
+ * (the "License"); you may not use this file except in compliance with
+ * the License. You may obtain a copy of the License at
+ *
+ *http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Unless required by applicable law or agreed to in writing, software
+ * distributed under the License is distributed on an "AS IS" BASIS,
+ * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ * See the License for the specific language governing permissions and
+ * limitations under the License.
+ */
+
+package org.apache.spark.ml.evaluation
+
+import org.apache.spark.SparkContext
+import org.apache.spark.annotation.{Experimental, Since}
+import org.apache.spark.broadcast.Broadcast
+import org.apache.spark.ml.linalg.{BLAS, DenseVector, Vector, Vectors,
VectorUDT}
+import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
+import org.apache.spark.ml.param.shared.{HasFeaturesCol, HasPredictionCol}
+import org.apache.spark.ml.util.{DefaultParamsReadable,
DefaultParamsWritable, Identifiable, SchemaUtils}
+import org.apache.spark.sql.{DataFrame, Dataset}
+import org.apache.spark.sql.functions.{avg, col, udf}
+import org.apache.spark.sql.types.IntegerType
+
+/**
+ * :: Experimental ::
+ * Evaluator for clustering results.
+ * The metric computes the Silhouette measure
+ * using the squared Euclidean distance.
+ *
+ * The Silhouette is a measure for the validation
+ * of the consistency within clusters. It ranges
+ * between 1 and -1, where a value close to 1
+ * means that the points in a cluster are close
+ * to the other points in the same cluster and
+ * far from the points of the other clusters.
+ */
+@Experimental
+@Since("2.3.0")
+class ClusteringEvaluator @Since("2.3.0") (@Since("2.3.0") override val
uid: String)
+ extends Evaluator with HasPredictionCol with HasFeaturesCol with
DefaultParamsWritable {
+
+ @Since("2.3.0")
+ def this() = this(Identifiable.randomUID("cluEval"))
+
+ @Since("2.3.0")
+ override def copy(pMap: ParamMap): ClusteringEvaluator =
this.defaultCopy(pMap)
+
+ @Since("2.3.0")
+ override def isLargerBetter: Boolean = true
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setPredictionCol(value: String): this.type = set(predictionCol,
value)
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setFeaturesCol(value: String): this.type = set(featuresCol, value)
+
+ /**
+ * param for metric name in evaluation
+ * (supports `"silhouette"` (default))
+ * @group param
+ */
+ @Since("2.3.0")
+ val metricName: Param[String] = {
+val allowedParams = ParamValidators.inArray(Array("silhouette"))
+new Param(
+ this,
+ "metricName",
+ "metric name in evaluation (silhouette)",
+ allowedParams
+)
+ }
+
+ /** @group getParam */
+ @Since("2.3.0")
+ def getMetricName: String = $(metricName)
+
+ /** @group setParam */
+ @Since("2.3.0")
+ def setMetricName(value: String): this.type = set(metricName, value)
+
+ setDefault(metricName -> "silhouette")
+
+ @Since("2.3.0")
+ override def evaluate(dataset: Dataset[_]): Double = {
+SchemaUtils.checkColumnType(dataset.schema, $(featuresCol), new
VectorUDT)
+SchemaUtils.checkColumnType(dataset.schema, $(predictionCol),
IntegerType)
+
+$(metricName) match {
+ case "silhouette" =>
SquaredEuclideanSilhouette.computeSilhouetteScore(
+dataset,
+$(predictionCol),
+$(featuresCol)
+ )
--- End diff --
Reorg as:
```
$(metricName) match {
case "squaredSilhouette" =>
SquaredEuclideanSilhouette.computeSilhouetteScore(
dataset, $(predictionCol), $(featuresCol))
}
```
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19185: [Spark-21854] Added LogisticRegressionTrainingSum...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19185#discussion_r138047555 --- Diff: python/pyspark/ml/tests.py --- @@ -1478,6 +1478,40 @@ def test_logistic_regression_summary(self): sameSummary = model.evaluate(df) self.assertAlmostEqual(sameSummary.areaUnderROC, s.areaUnderROC) +def test_multiclass_logistic_regression_summary(self): +df = self.spark.createDataFrame([(1.0, 2.0, Vectors.dense(1.0)), + (0.0, 2.0, Vectors.sparse(1, [], [])), + (2.0, 2.0, Vectors.dense(2.0)), + (2.0, 2.0, Vectors.dense(1.9))], +["label", "weight", "features"]) +lr = LogisticRegression(maxIter=5, regParam=0.01, weightCol="weight", fitIntercept=False) +model = lr.fit(df) +self.assertTrue(model.hasSummary) +s = model.summary +# test that api is callable and returns expected types +self.assertTrue(isinstance(s.predictions, DataFrame)) +self.assertEqual(s.probabilityCol, "probability") +self.assertEqual(s.labelCol, "label") +self.assertEqual(s.featuresCol, "features") +self.assertEqual(s.predictionCol, "prediction") +objHist = s.objectiveHistory +self.assertTrue(isinstance(objHist, list) and isinstance(objHist[0], float)) +self.assertGreater(s.totalIterations, 0) +self.assertTrue(isinstance(s.labels, list)) +self.assertTrue(isinstance(s.truePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.falsePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.precisionByLabel, list)) +self.assertTrue(isinstance(s.recallByLabel, list)) +self.assertTrue(isinstance(s.fMeasureByLabel, list)) +self.assertAlmostEqual(s.accuracy, 0.75, 2) +self.assertAlmostEqual(s.weightedTruePositiveRate, 0.75, 2) +self.assertAlmostEqual(s.weightedFalsePositiveRate, 0.25, 2) +self.assertAlmostEqual(s.weightedRecall, 0.75, 2) +self.assertAlmostEqual(s.weightedPrecision, 0.583, 2) +self.assertAlmostEqual(s.weightedFMeasure, 0.65, 2) +# test evaluation (with training dataset) produces a summary with same values +# one check is enough to verify a summary is returned, Scala version runs full test --- End diff -- Please add test for evaluation like: ``` sameSummary = model.evaluate(df) self.assertAlmostEqual(sameSummary.accuracy, s.accuracy) ``` --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19185: [Spark-21854] Added LogisticRegressionTrainingSum...
Github user yanboliang commented on a diff in the pull request: https://github.com/apache/spark/pull/19185#discussion_r138048004 --- Diff: python/pyspark/ml/tests.py --- @@ -1478,6 +1478,40 @@ def test_logistic_regression_summary(self): sameSummary = model.evaluate(df) self.assertAlmostEqual(sameSummary.areaUnderROC, s.areaUnderROC) +def test_multiclass_logistic_regression_summary(self): +df = self.spark.createDataFrame([(1.0, 2.0, Vectors.dense(1.0)), + (0.0, 2.0, Vectors.sparse(1, [], [])), + (2.0, 2.0, Vectors.dense(2.0)), + (2.0, 2.0, Vectors.dense(1.9))], +["label", "weight", "features"]) +lr = LogisticRegression(maxIter=5, regParam=0.01, weightCol="weight", fitIntercept=False) +model = lr.fit(df) +self.assertTrue(model.hasSummary) +s = model.summary +# test that api is callable and returns expected types +self.assertTrue(isinstance(s.predictions, DataFrame)) +self.assertEqual(s.probabilityCol, "probability") +self.assertEqual(s.labelCol, "label") +self.assertEqual(s.featuresCol, "features") +self.assertEqual(s.predictionCol, "prediction") +objHist = s.objectiveHistory +self.assertTrue(isinstance(objHist, list) and isinstance(objHist[0], float)) +self.assertGreater(s.totalIterations, 0) +self.assertTrue(isinstance(s.labels, list)) +self.assertTrue(isinstance(s.truePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.falsePositiveRateByLabel, list)) +self.assertTrue(isinstance(s.precisionByLabel, list)) +self.assertTrue(isinstance(s.recallByLabel, list)) +self.assertTrue(isinstance(s.fMeasureByLabel, list)) +self.assertAlmostEqual(s.accuracy, 0.75, 2) +self.assertAlmostEqual(s.weightedTruePositiveRate, 0.75, 2) +self.assertAlmostEqual(s.weightedFalsePositiveRate, 0.25, 2) +self.assertAlmostEqual(s.weightedRecall, 0.75, 2) +self.assertAlmostEqual(s.weightedPrecision, 0.583, 2) +self.assertAlmostEqual(s.weightedFMeasure, 0.65, 2) --- End diff -- We need to add these check for the above ```test_logistic_regression_summary``` and rename it to ```test_binary_logistic_regression_summary```, since binary logistic regression summary has these variables as well. --- - To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19185: [Spark-21854] Added LogisticRegressionTrainingSum...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19185#discussion_r138046547
--- Diff: python/pyspark/ml/classification.py ---
@@ -603,6 +614,112 @@ def featuresCol(self):
"""
return self._call_java("featuresCol")
+@property
+@since("2.3.0")
+def labels(self):
+"""
+Returns the sequence of labels in ascending order. This order
matches the order used
+in metrics which are specified as arrays over labels, e.g.,
truePositiveRateByLabel.
+
+Note: In most cases, it will be values {0.0, 1.0, ...,
numClasses-1}, However, if the
+training set is missing a label, then all of the arrays over labels
+(e.g., from truePositiveRateByLabel) will be of length
numClasses-1 instead of the
+expected numClasses.
+"""
+return self._call_java("labels")
+
+@property
+@since("2.3.0")
+def truePositiveRateByLabel(self):
+"""
+Returns true positive rate for each label (category).
+"""
+return self._call_java("truePositiveRateByLabel")
+
+@property
+@since("2.3.0")
+def falsePositiveRateByLabel(self):
+"""
+Returns false positive rate for each label (category).
+"""
+return self._call_java("falsePositiveRateByLabel")
+
+@property
+@since("2.3.0")
+def precisionByLabel(self):
+"""
+Returns precision for each label (category).
+"""
+return self._call_java("precisionByLabel")
+
+@property
+@since("2.3.0")
+def recallByLabel(self):
+"""
+Returns recall for each label (category).
+"""
+return self._call_java("recallByLabel")
+
+@property
--- End diff --
Remove this annotation.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
[GitHub] spark pull request #19185: [Spark-21854] Added LogisticRegressionTrainingSum...
Github user yanboliang commented on a diff in the pull request:
https://github.com/apache/spark/pull/19185#discussion_r138047016
--- Diff: python/pyspark/ml/classification.py ---
@@ -603,6 +614,112 @@ def featuresCol(self):
"""
return self._call_java("featuresCol")
+@property
+@since("2.3.0")
+def labels(self):
+"""
+Returns the sequence of labels in ascending order. This order
matches the order used
+in metrics which are specified as arrays over labels, e.g.,
truePositiveRateByLabel.
+
+Note: In most cases, it will be values {0.0, 1.0, ...,
numClasses-1}, However, if the
+training set is missing a label, then all of the arrays over labels
+(e.g., from truePositiveRateByLabel) will be of length
numClasses-1 instead of the
+expected numClasses.
+"""
+return self._call_java("labels")
+
+@property
+@since("2.3.0")
+def truePositiveRateByLabel(self):
+"""
+Returns true positive rate for each label (category).
+"""
+return self._call_java("truePositiveRateByLabel")
+
+@property
+@since("2.3.0")
+def falsePositiveRateByLabel(self):
+"""
+Returns false positive rate for each label (category).
+"""
+return self._call_java("falsePositiveRateByLabel")
+
+@property
+@since("2.3.0")
+def precisionByLabel(self):
+"""
+Returns precision for each label (category).
+"""
+return self._call_java("precisionByLabel")
+
+@property
+@since("2.3.0")
+def recallByLabel(self):
+"""
+Returns recall for each label (category).
+"""
+return self._call_java("recallByLabel")
+
+@property
+@since("2.3.0")
+def fMeasureByLabel(self, beta=1.0):
+"""
+Returns f-measure for each label (category).
+"""
+return self._call_java("fMeasureByLabel", beta)
+
+@property
+@since("2.3.0")
+def accuracy(self):
+"""
+Returns accuracy.
+(equals to the total number of correctly classified instances
+out of the total number of instances.)
+"""
+return self._call_java("accuracy")
+
+@property
+@since("2.3.0")
+def weightedTruePositiveRate(self):
+"""
+Returns weighted true positive rate.
+(equals to precision, recall and f-measure)
+"""
+return self._call_java("weightedTruePositiveRate")
+
+@property
+@since("2.3.0")
+def weightedFalsePositiveRate(self):
+"""
+Returns weighted false positive rate.
+"""
+return self._call_java("weightedFalsePositiveRate")
+
+@property
+@since("2.3.0")
+def weightedRecall(self):
+"""
+Returns weighted averaged recall.
+(equals to precision, recall and f-measure)
+"""
+return self._call_java("weightedRecall")
+
+@property
+@since("2.3.0")
+def weightedPrecision(self):
+"""
+Returns weighted averaged precision.
+"""
+return self._call_java("weightedPrecision")
+
+@property
--- End diff --
Remove this annotation.
---
-
To unsubscribe, e-mail: reviews-unsubscr...@spark.apache.org
For additional commands, e-mail: reviews-h...@spark.apache.org
